Have you ever wondered how your body creates new cells? Or how genetic information is passed down from one generation to the next? The answer lies in a fascinating process called DNA replication. In this article, we’ll dive deep into the world of eukaryotic DNA replication, exploring its intricacies, importance, and the cutting-edge research surrounding it.
What is Eukaryotic DNA Replication?
- Eukaryotic DNA replication is a crucial process that ensures the faithful duplication of genetic material in eukaryotic cells. This article provides an in-depth understanding of eukaryotic DNA replication, highlighting its significance and key molecular mechanisms.
- During DNA replication, DNA polymerases synthesize a complementary DNA strand based on the original template strand. The process begins with DNA helicases unwinding the double-stranded DNA, forming a replication fork that contains two single-stranded templates. This replication fork is where the replication machinery, known as the replisome, coordinates various proteins to carry out the enzymatic functions necessary for DNA synthesis. While the fundamental enzymatic functions are similar in prokaryotes and eukaryotes, eukaryotic DNA replication involves a larger and more complex set of proteins.
- The replisome is responsible for copying the entire genomic DNA in each dividing cell. This precise replication process ensures the accurate transmission of genetic information from parent cells to daughter cells, making it essential for all organisms. The cell cycle is carefully regulated to ensure error-free DNA replication. In the G1 phase, the regulatory processes for DNA replication initiation are initiated. The majority of DNA synthesis occurs during the S phase, where the entire genome is unwound and duplicated to form two daughter copies. In the G2 phase, any DNA damage or replication errors are corrected. Finally, during mitosis or M phase, one copy of the genomes is segregated into each daughter cell.
- Eukaryotic DNA replication follows the process of semiconservative replication, which is conserved from prokaryotes. This means that each parental DNA strand acts as a template for the synthesis of a new DNA strand. The replication fork serves as the site for DNA replication, where the DNA helix is unwound, exposing unpaired DNA nucleotides. These nucleotides are recognized and paired with complementary free nucleotides, leading to the formation of double-stranded DNA.
- It is important to note that eukaryotic DNA replication differs from prokaryotic replication due to the larger size and complexity of eukaryotic genomes. Eukaryotic replication initiation involves multiple initiator proteins, unlike prokaryotic genomes with specific replication sites. However, the overall process remains the same, although different enzymes are involved. The rate of eukaryotic DNA replication is slower compared to prokaryotes, typically around 100 nucleotides per second.
- In summary, eukaryotic DNA replication is a highly regulated and essential process that ensures the accurate duplication of genetic material. It involves the coordinated actions of various enzymes and proteins to synthesize a complementary DNA strand based on the original template. Understanding the mechanisms of eukaryotic DNA replication is crucial for comprehending how genetic information is inherited and passed on from one generation to the next.
Features of Eukaryotic DNA Replication
Eukaryotic DNA replication is a complex and precisely regulated process essential for cell division. This process ensures that each daughter cell receives an accurate copy of the genetic material. Here are the key features of eukaryotic DNA replication, explained step-by-step:
- Multiple Origins of Replication:
- Bi-directional Replication: Replication in eukaryotes begins at multiple origins of replication along the DNA molecule. These origins, known as Ori C, allow the process to proceed simultaneously at various points, ensuring efficient replication of the large eukaryotic genomes.
- Initiation: Initiation occurs at many chromosomal origins, which are recognized by specific initiator proteins that recruit the replication machinery.
- Semi-conservative Replication:
- Mechanism: Eukaryotic DNA replication follows a semi-conservative method. Each newly formed DNA molecule consists of one parental strand and one newly synthesized daughter strand. This ensures genetic continuity across generations.
- S Phase Specificity:
- Timing: DNA replication is restricted to the S phase (synthesis phase) of the cell cycle. This temporal regulation ensures that the entire genome is duplicated once per cell cycle, preventing re-replication.
- Nuclear Location:
- Site of Replication: The replication process takes place within the cell nucleus, where the eukaryotic DNA is housed. This compartmentalization allows for precise control and coordination of replication with other nuclear processes.
- Directional Synthesis:
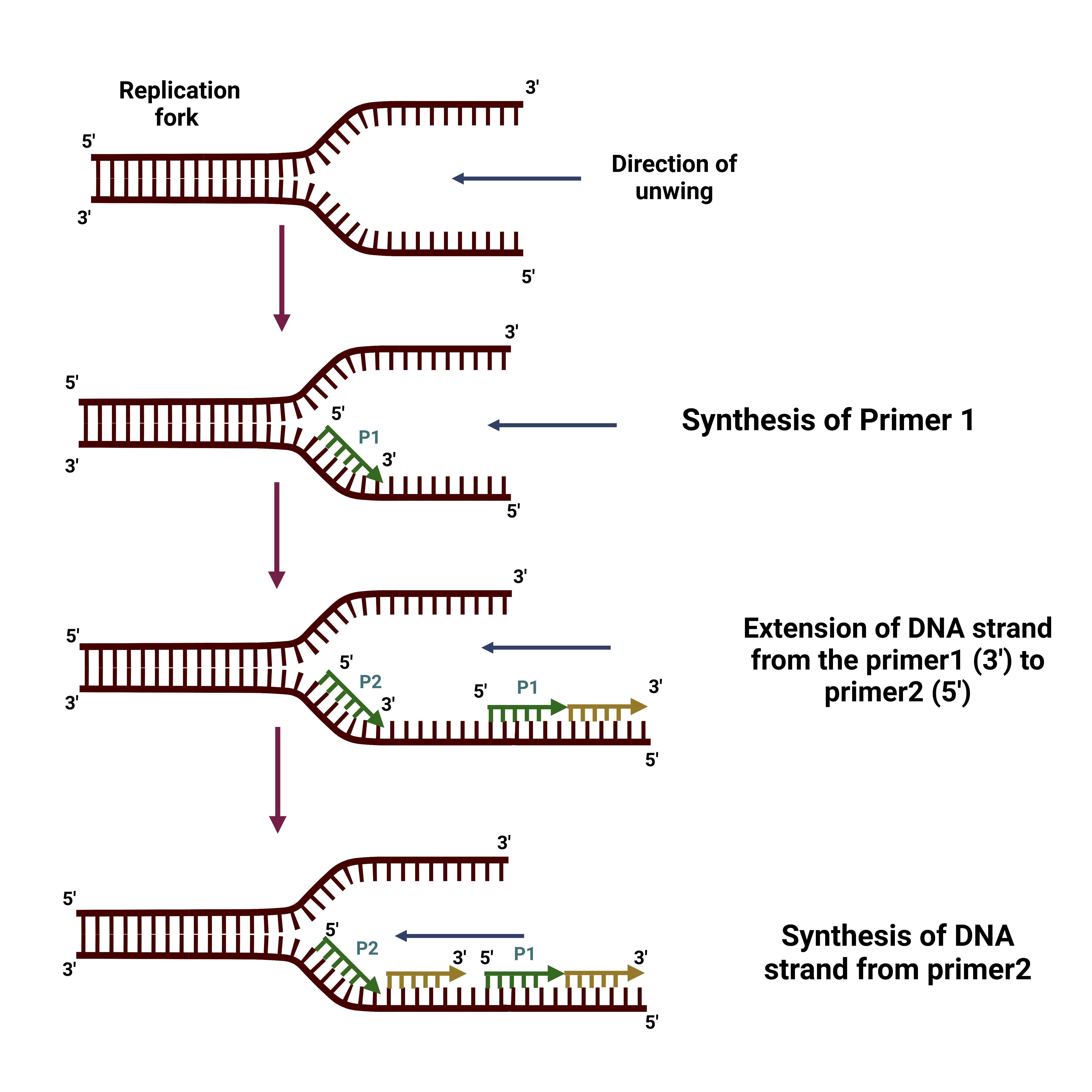
- 5′ to 3′ Direction: DNA synthesis occurs exclusively in the 5′ to 3′ direction. This directional constraint necessitates distinct mechanisms for replicating the two strands of the DNA helix.
- Leading and Lagging Strands:
- Continuous and Discontinuous Synthesis: The two strands of DNA are synthesized in different directions. The leading strand is synthesized continuously in the direction of the replication fork movement. Conversely, the lagging strand is synthesized discontinuously, forming short DNA segments known as Okazaki fragments.
- Formation of Okazaki Fragments:
- Lagging Strand Synthesis: The lagging strand is synthesized in short, separate segments called Okazaki fragments. These fragments are later joined together by DNA ligase to form a continuous strand, ensuring complete replication of the lagging strand.
- Role of DNA Polymerases:
- Polymerase Diversity: Eukaryotic cells possess multiple types of DNA polymerases involved in replication, each with specialized functions. The five primary polymerases are:
- Pol α (Alpha): Initiates DNA synthesis by adding RNA primers.
- Pol δ (Delta): Extends the RNA-primed DNA on the lagging strand.
- Pol ε (Epsilon): Primarily responsible for leading strand synthesis.
- Pol γ (Gamma): Replicates mitochondrial DNA.
- Pol β (Beta): Involved in DNA repair processes.
- Polymerase Diversity: Eukaryotic cells possess multiple types of DNA polymerases involved in replication, each with specialized functions. The five primary polymerases are:
These features collectively ensure that eukaryotic DNA replication is highly accurate and efficiently coordinated with the cell cycle.
What is DNA Polymerases?
Eukaryotic cells utilize a diverse array of DNA polymerases to manage DNA replication and repair, ensuring genomic stability and accurate genetic inheritance. Here, we explore the specific roles and functions of the primary DNA polymerases in eukaryotic cells.
- DNA Polymerases α, β, γ, δ, and ε:
- Pol α (Alpha):
- Function: Initiates DNA synthesis on both the leading and lagging strands by adding RNA primers.
- Primase Subunit: Contains a primase subunit that synthesizes short RNA primers necessary for DNA polymerase δ to extend.
- Lagging Strand Synthesis: Works in conjunction with Pol δ to synthesize the lagging strand via Okazaki fragments.
- Pol β (Beta):
- Function: Involved primarily in DNA repair processes.
- Role: Repairs short patch base excision, an essential process to correct DNA damage and maintain genetic integrity.
- Pol γ (Gamma):
- Function: Responsible for replicating mitochondrial DNA.
- Mitochondrial Replication: Ensures the faithful duplication of the mitochondrial genome, which is crucial for mitochondrial function and energy production.
- Pol δ (Delta):
- Function: Extends RNA primers initiated by Pol α and synthesizes the leading strand.
- Lagging Strand Synthesis: Collaborates with Pol α in synthesizing the lagging strand by filling in gaps between Okazaki fragments.
- Pol ε (Epsilon):
- Function: Primarily involved in the synthesis of the leading strand.
- DNA Repair: Also plays a significant role in DNA repair mechanisms, ensuring the correction of errors during replication.
- Pol α (Alpha):
- Additional Key Enzymes:
- Telomerase:
- Function: A specialized DNA polymerase that replicates the ends of chromosomes (telomeres).
- Integral RNA Primer: Contains its own RNA template to extend telomeres, compensating for the inability of conventional DNA polymerases to fully replicate the ends of linear chromosomes.
- DNA Topoisomerase I:
- Function: Relaxes the DNA helix by creating a transient nick in one of the DNA strands.
- Role: Alleviates the torsional strain that builds up ahead of the replication fork, ensuring smooth progression of the replication machinery.
- DNA Topoisomerase II:
- Function: Relieves the strain on the DNA helix by forming supercoils and creating transient nicks in both DNA strands.
- Role: Essential for untangling intertwined DNA molecules, thereby facilitating proper chromosome segregation during cell division.
- DNA Ligase:
- Function: Forms 3′-5′ phosphodiester bonds between adjacent DNA fragments.
- Role: Joins Okazaki fragments on the lagging strand, ensuring the creation of a continuous DNA strand and completing the replication process.
- Telomerase:
Eukaryotic DNA replication protein list
| Protein | Function in Eukaryotic DNA Replication |
|---|---|
| AND1 | Loads DNA polymerase α onto chromatin together with CMG complex on the lagging strand. Also known as Ctf4 in budding yeast. |
| Cdc45 | Required for initiation and elongation steps of DNA replication. A part of the Mcm2-7 helicase complex. Required after pre-RC step for loading of various proteins for initiation and elongation. |
| Cdc45-Mcm-GINS (CMG) complex | Functional DNA helicase in eukaryotic cells |
| Cdc6 | Required for assembly of Mcm2-7 complex at ORC, in conjunction with Cdt1. |
| Cdc7-Dbf4 kinase or Dbf4-dependent kinase (DDK) | Protein kinase required for initiation of DNA replication, probably through phosphorylation of the minichromosome maintenance proteins. |
| Cdt1 | Loads Mcm2-7 complex on DNA at ORC in pre-RC/licensing step. Inhibited in metazoans by geminin. |
| Claspin | Couples leading-strand synthesis with the CMG complex helicase activity. Works with Mrc1. |
| Ctf4 | Loads DNA polymerase α onto chromatin together with CMG complex on the lagging strand. Homolog in metazoans is known as AND-1. |
| Cyclin-dependent kinase (CDK) | Cyclin-dependent protein kinase required for initiation of replication and for other subsequent steps. |
| Dna2 | 5′ flap endonuclease and helicase involved in processing Okazaki fragments. |
| DNA ligase I | Joins Okazaki fragments during DNA replication. Ligase activity also needed for DNA repair and recombination. |
| DNA polymerase α (Pol α) | Contains primase activity that is necessary to initiate DNA synthesis on both leading and lagging strands. |
| DNA polymerase δ (Pol δ) | Required to complete synthesis of Okazaki fragments on the lagging strand that have been started by DNA polymerase α. |
| DNA polymerase ε (Pol ε) | The leading strand polymerase. Synthesizes DNA at the replication fork. Binds early at origins via Dbp11 and needed to load DNA polymerase α. |
| Dpb11 | DNA replication initiation protein. Loads DNA polymerase ε onto pre-replication complexes at origins. |
| Fen1 | 5′ flap endonuclease involved in processing Okazaki fragments. |
| Geminin | Protein found in metazoans and absent from yeasts. Binds to and inactivates Cdt1, thereby regulating pre-replicative/initiation complex formation. Also suggested to promote pre-RC formation by binding and thus preventing Cdt1 degradation. |
| GINS | Tetrameric complex composed of Sld5, Psf1, Psf2, Psf3. Associates with pre-replicative complex around the time of initiation and moves with replication forks during the elongation step. Required for the elongation stage of DNA replication and may be part of the Mcm helicase complex. |
| Minichromosome maintenance proteins (Mcm) | Six different proteins of the AAA+ ATPase family that form a hexamer in solution. This hexamer is recruited and loaded by ORC, Cdc6, and Cdt1 and forms a double hexamer that is topologically linked around DNA to form a salt-resistant pre-replicative complex. On replication initiation, Mcm2-7 moves away from ORC with the replication fork. |
| Mcm10 | Required for initiation and elongation stages of DNA replication. Implicated in chromatin binding of Cdc45 and DNA polymerase α. Also required for stability of DNA polymerase α catalytic subunit in the budding yeast S. cerevisiae. |
| Mrc1 | Couples leading-strand synthesis with the CMG complex helicase activity. Metazoan homolog is known as Claspin. |
| Origin recognition complex (ORC) | Heterohexameric complex composed of Orc1–Orc6 proteins. Binds to DNA and assembles Mcm2-7 complex onto chromatin together with Cdc6 and Cdt1. |
| Proliferating cell nuclear antigen (PCNA) | Trimeric protein with a ring-shaped structure that encloses DNA, preventing dissociation of DNA polymerase. Acts as a sliding clamp for polymerases δ and ε, thereby improving processivity of replicative polymerases. |
| Replication factor C (RFC) | Loads PCNA on primed templates and is involved in the switch between DNA polymerase α and the replicative polymerases δ and ε. |
| Replication fork barriers (RFBs) | Bound by RFB proteins in various locations throughout the genome. Are able to terminate or pause replication forks, stopping the progression of the replisome. |
| Replication protein A (RPA) | Heterotrimeric single-stranded binding protein. Stabilizes single-stranded DNA at the replication fork. |
| RNase H | Ribonuclease which digests RNA hybridized to DNA. Involved in Okazaki fragment processing. |
| Sld2 | Functions in the initiation of replication. Key substrate of CDK, phosphorylation promotes interaction with Dpb11. Required for initiation of replication. |
| Sld3 | Functions in the initiation of replication. Key substrate of CDK, phosphorylation promotes interaction with Dpb11. Required for initiation of replication. |
| Telomerase | A ribonucleoprotein that adds DNA sequence “TTAGGG” repeats to the 3′ end of DNA strands in telomeres. |
| Topoisomerases | Regulate the overwinding or underwinding of DNA. |
Overview of Eukaryotic DNA Replication Steps
Eukaryotic DNA replication is a highly orchestrated and complex process, essential for cell division and genetic continuity. This process involves multiple steps and components, ensuring the accurate duplication of the genome. Below is a detailed, step-by-step explanation of eukaryotic DNA replication:
- Initiation at Multiple Origins:
- Origins of Replication: Replication begins at multiple origins along each linear DNA molecule, approximately every 30–300 kilobases, depending on the species and tissue type.
- Replication Bubbles: At each origin, a replication bubble forms, with two replication forks moving bi-directionally. The DNA replicated under the control of a single origin is termed a replicon.
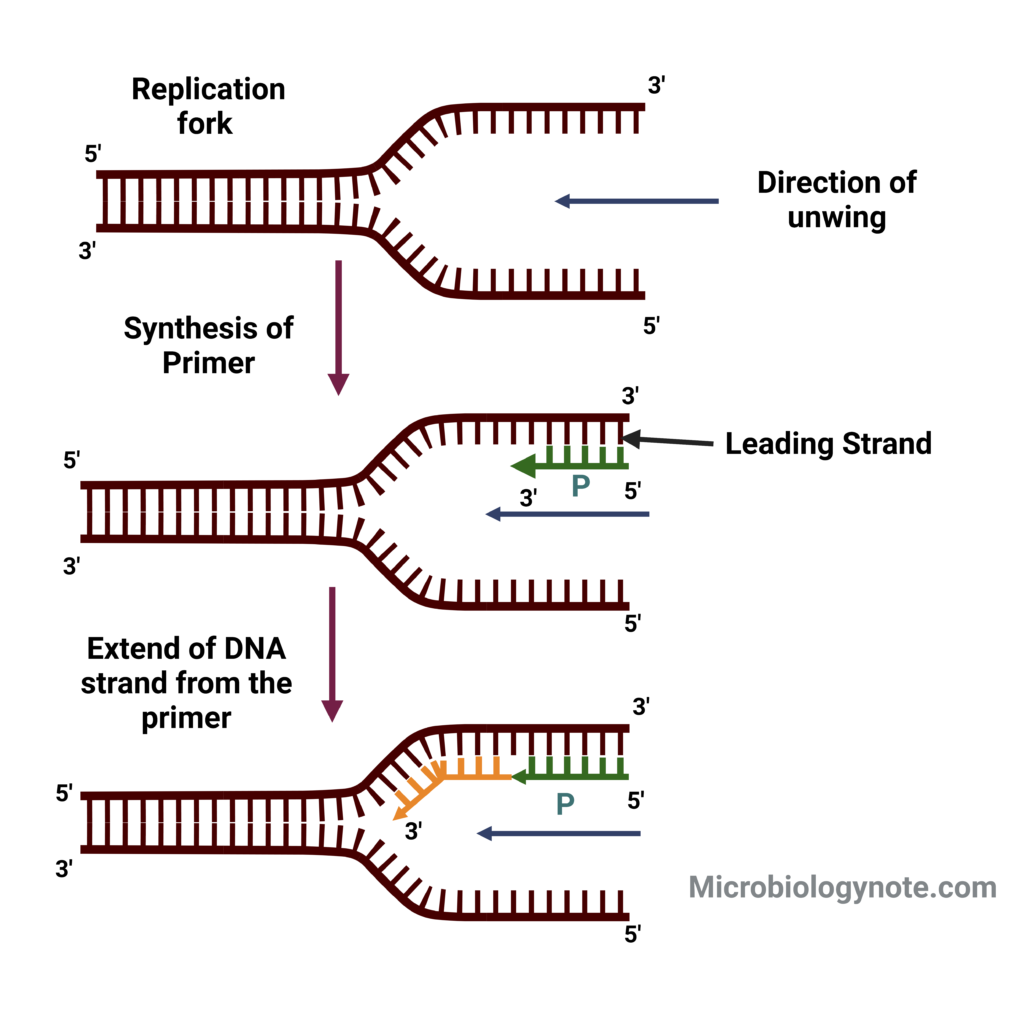
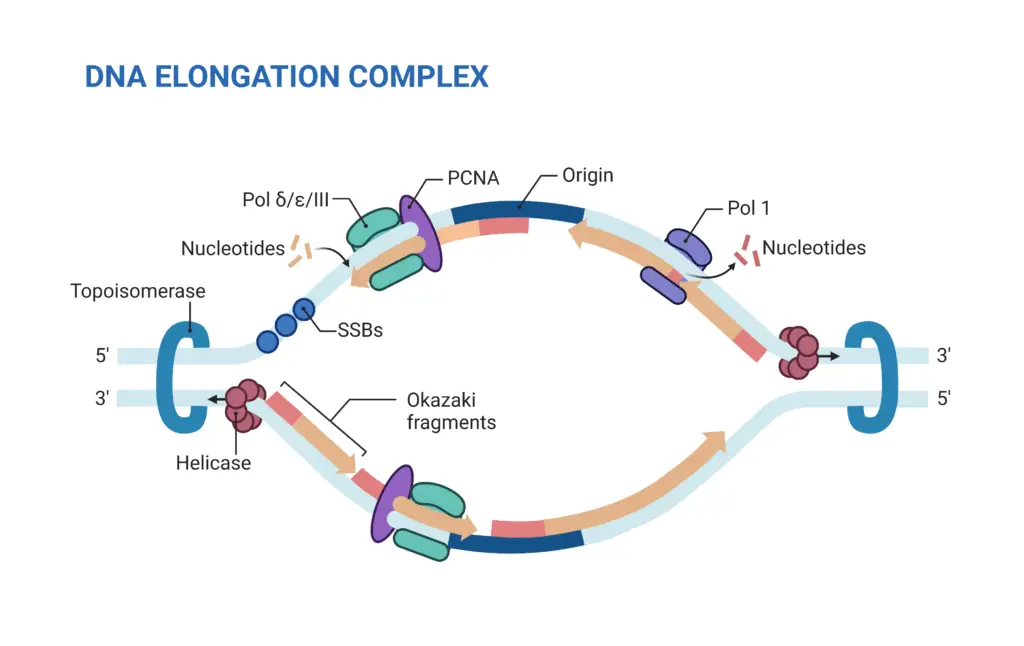
- Formation and Stabilization of the Replication Fork:
- Helicase Activity: Enzymes called helicases unwind the double helix at the origin, creating two replication forks.
- SSB Proteins: Single-strand binding proteins (SSBs) stabilize the unwound DNA, preventing it from re-annealing.
- Topoisomerases: DNA topoisomerases alleviate the torsional strain generated ahead of the replication fork by creating transient nicks in the DNA strands.
- Primer Synthesis:
- DNA Polymerase α: This enzyme, carrying a primase subunit, synthesizes RNA primers complementary to the DNA template. These primers are essential for initiating DNA synthesis.
- Elongation of the Leading and Lagging Strands:
- Leading Strand Synthesis:
- DNA Polymerase δ: The leading strand is synthesized continuously in the 5′ to 3′ direction by DNA polymerase δ.
- Lagging Strand Synthesis:
- Okazaki Fragments: The lagging strand is synthesized discontinuously in short segments called Okazaki fragments. DNA polymerase α initiates these fragments with an RNA primer, then extends it with a short DNA segment.
- DNA Polymerase δ: After the initial synthesis by DNA polymerase α, DNA polymerase δ synthesizes the remainder of the Okazaki fragment.
- Leading Strand Synthesis:
- Joining of Okazaki Fragments:
- DNA Ligase: This enzyme seals the nicks between Okazaki fragments and around RNA primers, forming a continuous DNA strand. It achieves this by creating phosphodiester bonds between adjacent DNA fragments.
- Completion of DNA Synthesis:
- Replication Bubble Merging: DNA synthesis proceeds until adjacent replication bubbles merge, resulting in the complete replication of the DNA molecule.
The intricacies of eukaryotic DNA replication, including the roles of various enzymes and the sequential synthesis of leading and lagging strands, underscore the precision of cellular replication mechanisms. This process ensures the faithful transmission of genetic information from one generation to the next, maintaining the integrity of the genome across cellular divisions.
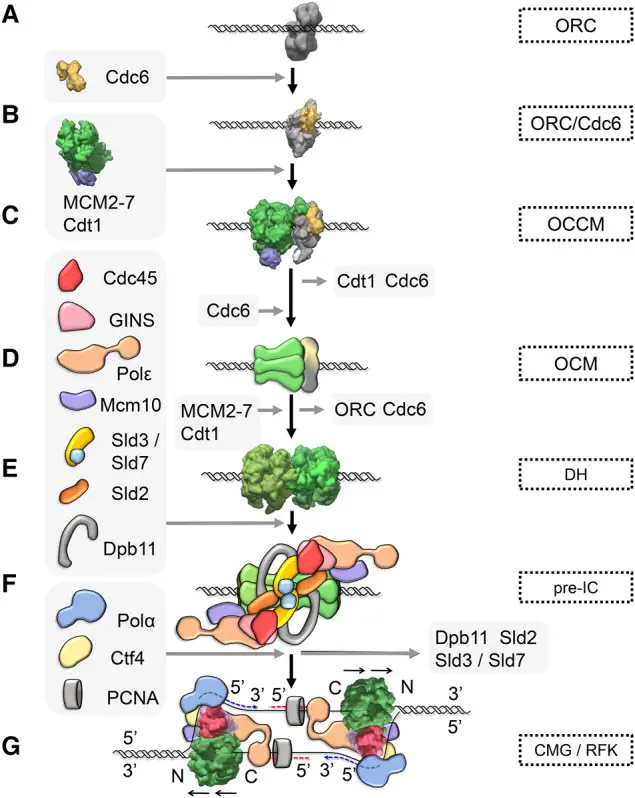
Initiation of Eukaryotic DNA replication
The initiation of eukaryotic DNA replication is a meticulously regulated process that ensures accurate duplication of the genome. This process involves several key steps, each facilitated by specific proteins and complexes. Here is a detailed, step-by-step explanation of the initiation of eukaryotic DNA replication:
- Origin of Replication:
- Definition and Distribution: Replication begins at specific locations in the genome called origins of replication. Eukaryotic genomes, being significantly larger than prokaryotic ones, have numerous origins to expedite the replication process.
- Origin Recognition Complex (ORC): The origin recognition complex (ORC) binds to the origin sequences. ORC consists of six subunits organized in a C-shaped structure that encircles the DNA at the origin, serving as a platform for the recruitment of other replication proteins.
- Formation of the ORC/Cdc6/DNA Complex:
- Cdc6 Binding: During the late M phase, the cell division cycle 6 (Cdc6) protein binds to the ORC/DNA complex. This binding requires ATP, forming the ORC/Cdc6/DNA complex.
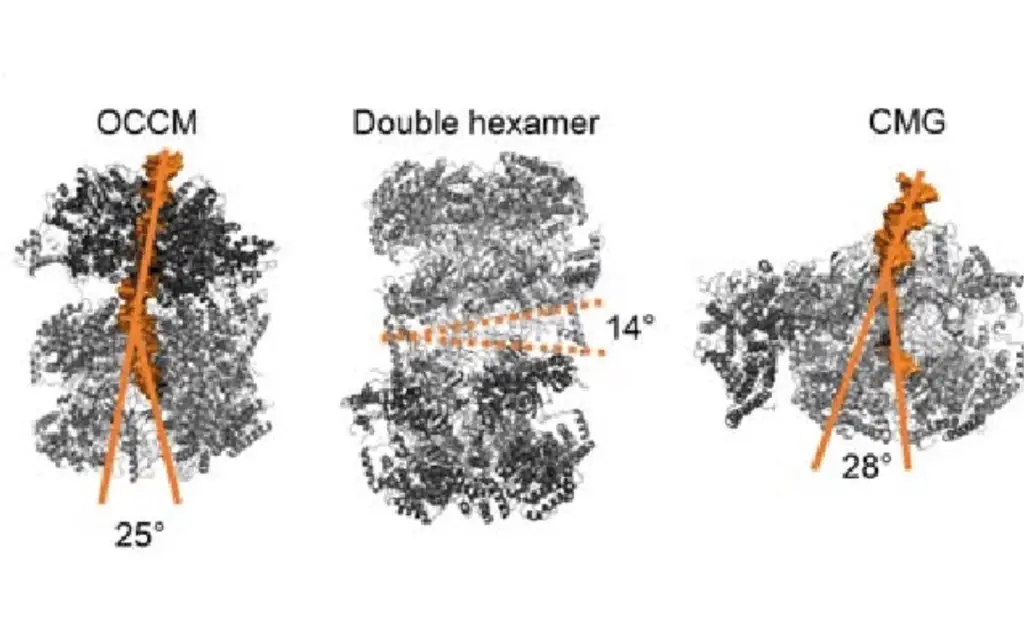
- Formation of the OCCM Complex:
- Cdt1 and MCM2-7 Loading: The heptamer Cdt1/MCM2-7 binds to the ORC/Cdc6 complex, leading to the loading of the minichromosome maintenance (MCM) 2-7 helicase onto the DNA. MCM2-7 is crucial for unwinding the DNA helix.
- Formation of the OM Intermediate:
- Release of Cdc6 and Cdt1: ATP hydrolysis facilitates the release of Cdc6 and Cdt1, resulting in the formation of the ORC/MCM2-7 (OM) intermediate.
- Formation of the OCM Complex:
- Binding of a Second MCM2-7 Hexamer: Another Cdc6 molecule binds to the OM intermediate, allowing the attachment of a second MCM2-7 hexamer. This leads to the formation of the ORC/Cdc6/MCM2-7 (OCM) complex.
- Formation of the Double Hexamer (DH):
- Alignment and Release: As the cell progresses through the G1 phase, a second MCM2-7 hexamer is recruited, aligning head-to-head with the first, forming a double hexamer (DH). Cdc6, ORC, and Cdt1 are released during this process.
- Formation of the Pre-Replication Complex (pre-RC):
- DNA Encirclement: The DH complex encircles the double-stranded DNA, marking the formation of the pre-replication complex (pre-RC). This step is also referred to as DNA licensing.
- Formation of the Pre-Initiation Complex (pre-IC):
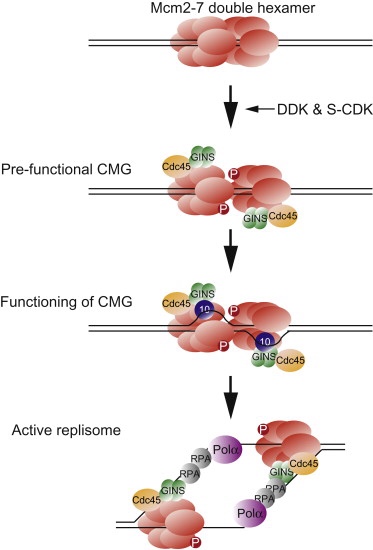
- Phosphorylation and Activation: During the onset of the S phase, the kinase DDK phosphorylates the MCM2-7 DHs, activating them. This phosphorylation event leads to the assembly of additional proteins to form the pre-initiation complex (pre-IC).
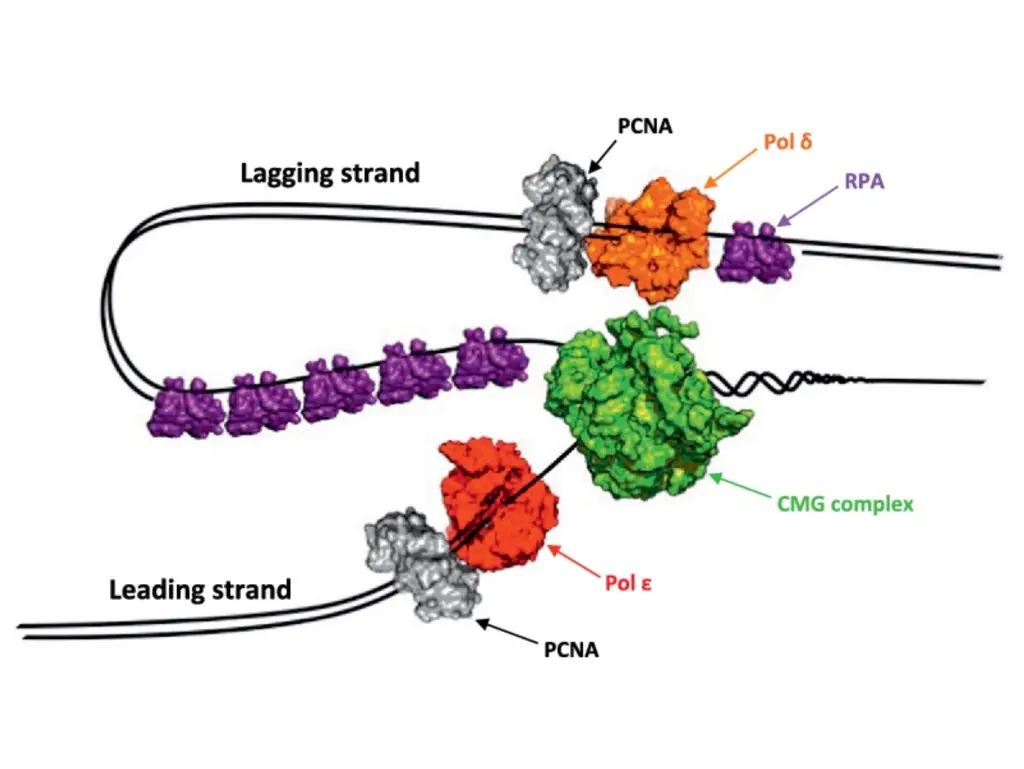
- Formation of the CMG Complex:
- Protein Recruitment: The phosphorylated MCM2-7 complex creates a binding site for the Sld3/Sld7 complex and Cdc45. Cyclin-dependent kinase (CDK) phosphorylates Sld2 and Sld3, which then interact with Dpb11, aiding in the recruitment of Cdc45 and GINS.
- Assembly of CMG Complex: The combined action of Sld2, Sld3, and Dpb11 facilitates the formation of the Cdc45/MCM2-7/GINS (CMG) complex, which is essential for the helicase activity at the replication fork.
- Loading of DNA Polymerase:
- Replication Protein A (RPA): RPA stabilizes the single-stranded DNA as it is unwound. Pol ε is then loaded onto the helicase with the help of Ctf4.
- Pol α-Primase Complex: Pol α, along with DNA primase, forms a complex responsible for synthesizing RNA primers and initiating DNA synthesis.
- Synthesis of Leading and Lagging Strands:
- Leading Strand: DNA polymerase ε continues synthesizing the leading strand continuously in the 5′ to 3′ direction.
- Lagging Strand: DNA polymerase δ synthesizes the lagging strand discontinuously through the formation of Okazaki fragments.
These steps ensure that the initiation of eukaryotic DNA replication is precise, controlled, and efficient, laying the groundwork for accurate and complete genome duplication.
Elongation of Eukaryotic DNA replication
Elongation during eukaryotic DNA replication is a highly coordinated process, involving the extension of the DNA strands by various polymerases and the formation of Okazaki fragments on the lagging strand. Here are the detailed steps involved:
- Initiation of DNA Synthesis:
- DNA Polymerase α-Primase Complex: The process begins with the DNA polymerase α-primase complex. The primase subunit synthesizes short RNA primers (7-12 nucleotides), which are then extended by the polymerase domain with DNA bases (approximately 20-25 nucleotides).
- Polymerase Switching: Replication factor C (RFC) initiates polymerase switching, replacing DNA polymerase α with other polymerases to elongate the DNA strands.
- Leading Strand Synthesis:
- PCNA Loading: After DNA polymerase α synthesizes the RNA primer and initial DNA bases, RFC facilitates the assembly of proliferating cell nuclear antigen (PCNA) at the primer terminus. PCNA acts as a DNA clamp for polymerases.
- Pol ε Activity: DNA polymerase ε is primarily responsible for continuous synthesis of the leading strand. The process is straightforward, requiring only a single round of primer synthesis.
- Lagging Strand Synthesis:
- Okazaki Fragments: The lagging strand is synthesized in short fragments called Okazaki fragments, each about 200 bases long in eukaryotes. DNA polymerase α initiates these fragments by adding DNA bases to an RNA primer.
- Pol δ Activity: DNA polymerase δ extends these fragments. RFC-triggered polymerase switching ensures that PCNA holds the polymerase in place. When Pol δ reaches the previous RNA primer, it dissociates, signaling completion of that fragment.
- Primer Removal and Gap Filling: RNase H1 degrades RNA primers, leaving a single ribonucleotide. Flap endonuclease 1 (FEN1) removes this nucleotide, and DNA polymerase δ fills the gaps. DNA ligase I then seals the nicks between fragments, creating a continuous strand.
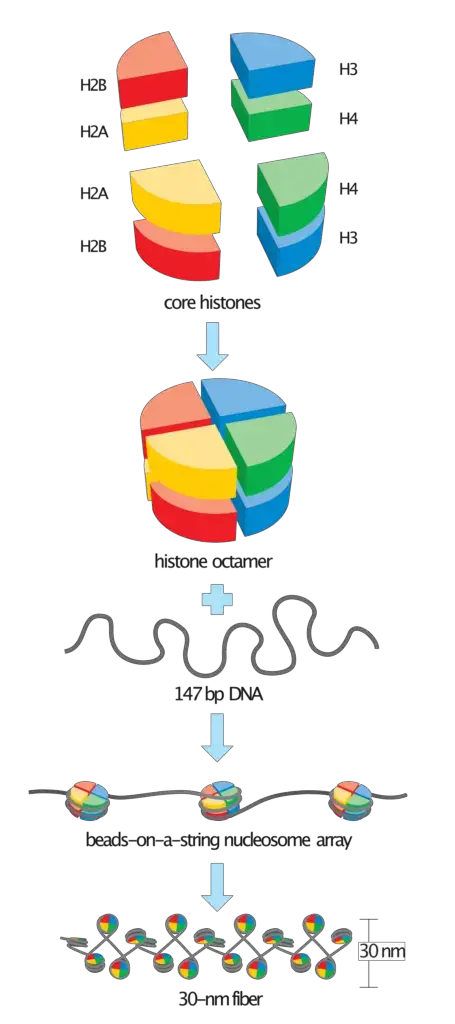
- Nucleosome Assembly:
- Nucleosome Disassembly: DNA replication disrupts nucleosome structures in front of the replication fork, allowing replication proteins to access the DNA.
- Nucleosome-Free Regions: As replication proceeds, nucleosome-free regions form, extending around 225 base pairs (bp) on the leading strand and 285 bp on the lagging strand.
- Histone Reassembly: Parental nucleosomes randomly bind to the daughter DNA strands. New nucleosome components are synthesized and assembled onto the daughter strands, ensuring each strand receives the necessary histone proteins.
- Completion of Nucleosome Formation: After replication, complete nucleosomes containing histone H1 are assembled, stabilizing the newly synthesized DNA.
Termination of Eukaryotic DNA Replication
Termination of eukaryotic DNA replication is a complex, coordinated process essential for ensuring that each daughter cell receives an accurate copy of the genome. Here are the detailed steps involved:
Involving Two Adjacent Replication Forks
Termination in eukaryotic cells requires the merger of two neighboring replication forks due to the multiple origins of replication. This process includes the following stages:
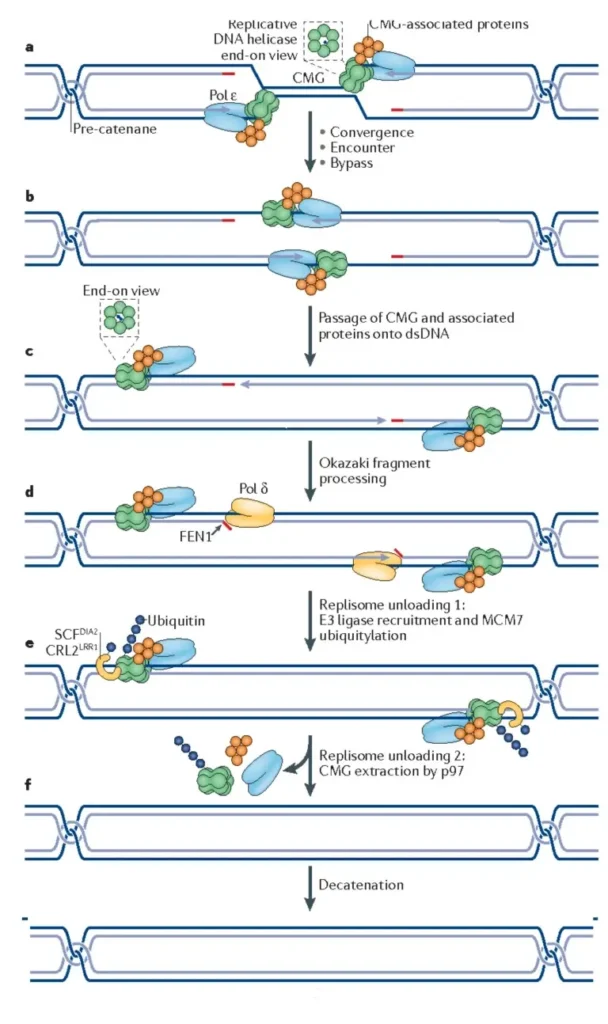
- Dissolution:
- Unwinding: The CMG helicase complexes (comprising Cdc45, Mcm2-7, and GINS) move towards each other, unwinding the DNA segment between the two replication forks.
- Okazaki Fragment Processing: DNA polymerase δ and the elongation factor FEN1 collaborate to process the final Okazaki fragment, ensuring complete replication.
- Ligation:
- Strand Joining: The newly synthesized DNA strands, approaching from opposite directions, are ligated together, effectively closing any gaps in the daughter strands.
- Dissociation of Replisome:
- Replisome Disassembly: Following the merger of the two replication forks, the replisome complex is disassembled. This involves the polyubiquitylation of Mcm7 and the action of the p97/VCP/Cdc48 segregase, ensuring the dismantling of the replication machinery.
- Decatenation:
- Separation of DNA Strands: Topoisomerase II is employed to resolve any entanglements or catenanes in the daughter strands, ensuring the two replicated DNA molecules are fully separated.
Involving the Ends of Chromosomes
Eukaryotic chromosomes are linear, and terminating DNA replication at the chromosome ends involves specific mechanisms to handle the unique challenges posed by these structures:
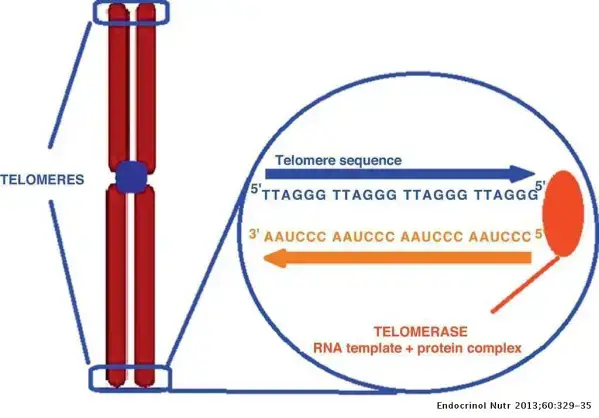
- Replication of Telomeres:
- Telomere Structure: Telomeres are repetitive DNA sequences (e.g., TTAGGG in humans) located at the ends of chromosomes, protecting them from degradation and preventing false signaling of DNA double-strand breaks.
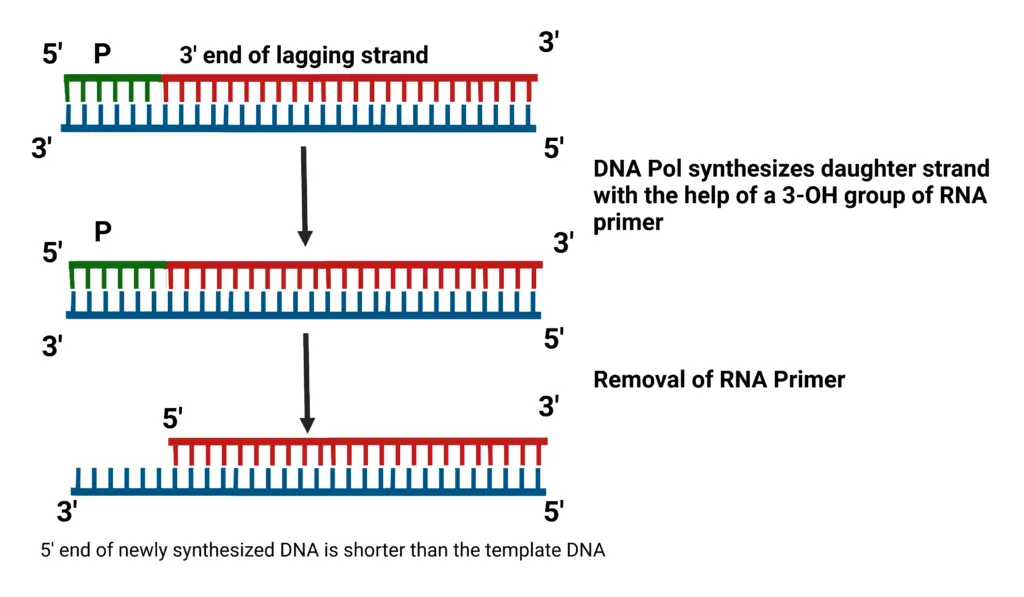
- Okazaki Fragment Replication: The 3′-OH group from an RNA primer initiates 5′-to-3′ replication during the synthesis of Okazaki fragments. However, upon removal of the RNA primer, a gap remains at the 5′ end, leading to progressive shortening of the chromosome with each replication cycle.
- Telomere Shortening and Cell Senescence:
- Normal Replication: In somatic cells, this shortening leads to replicative senescence or apoptosis once the telomeres become critically short, limiting the number of cell divisions.
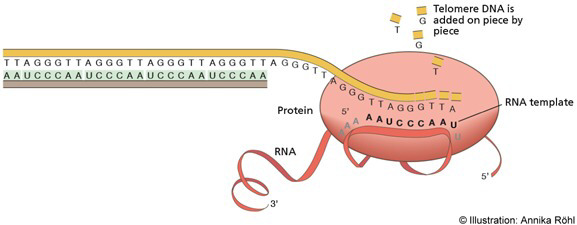
- Telomere Lengthening by Telomerase:
- Telomerase Function: In germline and cancer cells, the enzyme telomerase extends the 5′ end of the lagging strands, maintaining telomere length and contributing to cellular immortality.
- Telomerase Composition: Telomerase comprises a reverse transcriptase (TERT) and an RNA template (TER). In humans, the RNA template includes the sequence AUCCCAAUC, guiding the addition of telomeric repeats to the 3′ overhang of single-stranded DNA.
- Final Extension and Ligation: After telomerase has extended the 3′ end, DNA polymerase and DNA ligase complete the synthesis of the DNA strand, ensuring the telomeres are fully replicated and protected.
DNA Proofreading
DNA proofreading is a fundamental process ensuring the fidelity of DNA replication and the maintenance of genetic information. This mechanism is integral to eukaryotic cells and involves several key steps:
Steps in DNA Proofreading
- Polymerase Activity:
- Role of Polymerases: During DNA replication, DNA polymerases, particularly delta and epsilon polymerases, move along the template strand, adding complementary nucleotides to the growing daughter strand. These polymerases possess a built-in proofreading function through their 3′ to 5′ exonuclease activity.
- Error Detection: Occasionally, incorrect nucleotides are inserted during replication. The polymerases detect these errors, ensuring they do not perpetuate through the newly synthesized DNA.
- Proofreading Mechanism:
- Pause in Activity: Upon detecting an error, the polymerase temporarily halts its replication activity.
- Exonuclease Action: The 3′ to 5′ exonuclease activity enables the polymerase to backtrack along the newly synthesized strand. This backtracking is crucial for removing the incorrect nucleotide.
- Nucleotide Excision: The exonuclease activity excises the erroneous nucleotide from the growing strand, effectively correcting the mistake.
- Resume Elongation:
- Correct Nucleotide Addition: After removing the incorrect nucleotide, the polymerase resumes its elongation activity. It adds the correct nucleotide in place of the removed one, ensuring the DNA sequence is accurately replicated.
- Maintaining Integrity: This process maintains the genetic integrity by correcting errors as they occur, preventing mutations from propagating.
Excision Repair
Besides proofreading during replication, cells employ additional mechanisms to repair DNA damage caused by environmental factors, such as UV radiation. One of the critical repair mechanisms is excision repair.
- Damage Recognition:
- Types of Damage: Excision repair targets specific DNA lesions, such as pyrimidine dimers induced by UV rays or other mutated bases. These lesions distort the DNA structure, signaling the need for repair.
- Detection: Specialized proteins recognize the damaged DNA segment, initiating the repair process.
- Excision of Damaged DNA:
- Removal of Lesions: The damaged DNA segment is excised by nucleases, creating a gap in the DNA strand.
- Synthesis of New Strand: DNA polymerase fills the gap by synthesizing a new strand with the correct sequence, using the undamaged complementary strand as a template.
- Restoration:
- Ligation: DNA ligase seals the newly synthesized segment into the existing DNA strand, restoring the DNA’s continuity and integrity.
- Preservation of Genetic Code: This repair process preserves the accurate genetic code by fixing damaged regions and preventing mutations from becoming permanent.
Differences Between Prokaryotic DNA replication and Eukaryotic DNA replication
| Prokaryotic DNA replication | Eukaryotic DNA replication |
|---|---|
| Occurs inside the cytoplasm | Occurs inside the nucleus |
| Only one origin of replication per molecule of DNA | Have many origins of replication in each chromosome |
| Origin of replication is about 100-200 or more nucleotides in length | Each origin of replication is formed of about 150 nucleotides |
| Replication occurs at one point in each chromosome | Replication occurs at several points simultaneously in each chromosome |
| Only have one origin of replication | Has multiple origins of replication |
| Initiation is carried out by protein DnaA and DnaB | Initiation is carried out by the Origin Recognition Complex |
| Topoisomerase is needed | Topoisomerase is needed |
| Replication is very rapid | Replication is very slow |
Significance of Eukaryotic DNA Replication
DNA replication in eukaryotic cells is a vital process essential for cell growth, division, and the conservation of genetic information. This fundamental mechanism ensures the accurate transmission of genetic material and supports various cellular functions critical for life.
Key Functions of Eukaryotic DNA Replication
- Transmission of Genetic Information:
- Genome Duplication: Each eukaryotic cell contains a complete set of chromosomes representing the entire genome. During DNA replication, an identical copy of the genome is produced, ensuring that each daughter cell inherits a full set of genetic instructions.
- Generational Continuity: Accurate DNA replication is essential for passing genetic information from one generation to the next, maintaining species continuity.
- Cell Growth and Division:
- Cell Cycle Integration: Proper regulation of DNA replication is crucial for synchronized cell growth and division. The replication process must be precisely timed with the cell cycle to ensure genomic integrity.
- Mitotic Fidelity: During cell division, DNA replication ensures that each daughter cell receives the necessary genetic material, facilitating successful mitosis and cytokinesis.
- DNA Repair and Stability:
- Error Detection and Correction: DNA replication provides an opportunity for detecting and repairing DNA damage. This process is vital for maintaining genomic stability and preventing mutations.
- Genome Integrity: The replication machinery includes mechanisms to recognize and rectify errors, thereby preserving the accuracy of the genetic code.
- Genetic Recombination:
- Homologous Recombination: During replication, genetic material can be exchanged between homologous chromosomes through recombination. This exchange contributes to genetic diversity and is crucial for evolution and adaptation.
- Meiotic Processes: In sexually reproducing organisms, recombination during meiosis enhances genetic variability, promoting healthier offspring.
- Gene Expression and Regulation:
- Transcriptional Readiness: Replicated DNA is essential for gene expression, ensuring that the genetic instructions are available for transcription and translation.
- Regulatory Mechanisms: DNA replication influences the accessibility of genes, thereby playing a role in the regulation of gene expression.
- Development and Differentiation:
- Embryonic Development: During the development of multicellular organisms, DNA replication is meticulously regulated to support the formation of specialized cell types with distinct functions.
- Cell Differentiation: Proper replication ensures that differentiated cells receive the correct genetic information required for their specific roles within the organism.
Detailed and Sequential Explanation
- Initiation:
- Origin Recognition: Replication begins at specific sites called origins of replication. Proteins recognize these origins and initiate the unwinding of the DNA helix.
- Elongation:
- Polymerase Function: DNA polymerases add complementary nucleotides to the growing daughter strands. Leading and lagging strands are synthesized continuously and discontinuously, respectively.
- Proofreading: The 3′ to 5′ exonuclease activity of polymerases ensures that errors are corrected in real-time, maintaining replication fidelity.
- Termination:
- Fork Convergence: Replication forks converge and the newly synthesized strands are joined. Specific mechanisms ensure that replication concludes properly without leaving gaps or errors.
- Post-Replication Processes:
- Telomere Maintenance: Telomeres, the protective ends of chromosomes, are maintained to prevent genomic instability. Telomerase enzyme activity is crucial in this context.
- Chromatin Reassembly: After replication, newly synthesized DNA is packaged into chromatin, ensuring it is properly organized and functional for subsequent cellular processes.
FAQ
What is eukaryotic DNA replication?
Eukaryotic DNA replication is the process by which a eukaryotic cell duplicates its genetic material, specifically the DNA molecules that make up the chromosomes.
How does eukaryotic DNA replication differ from prokaryotic DNA replication?
Unlike prokaryotes, eukaryotic DNA replication occurs within the nucleus of the cell rather than in the cytoplasm. Eukaryotes also have multiple origins of replication per chromosome, while prokaryotes typically have only one.
What is the purpose of eukaryotic DNA replication?
The primary purpose of eukaryotic DNA replication is to ensure the accurate transmission of genetic information from one generation of cells to the next. It is essential for cell growth, division, and the maintenance of genetic integrity.
Which enzymes are involved in eukaryotic DNA replication?
Several enzymes play key roles in eukaryotic DNA replication, including DNA polymerases, helicases, topoisomerases, and ligases. These enzymes work together to unwind the DNA, synthesize new strands, and ensure proper DNA strand rejoining.
How does eukaryotic DNA replication ensure accuracy?
Eukaryotic DNA replication incorporates proofreading mechanisms to ensure accuracy. The DNA polymerases involved in replication possess exonuclease activity, allowing them to detect and remove incorrect nucleotides before continuing with replication.
What is the role of origins of replication in eukaryotic DNA replication?
Origins of replication are specific sites on the DNA where replication initiates. In eukaryotes, multiple origins of replication are present on each chromosome to facilitate efficient and timely replication.
How does eukaryotic DNA replication coordinate with other cellular processes?
Eukaryotic DNA replication is tightly regulated and coordinated with other cellular processes, such as cell cycle progression and DNA repair. This ensures that DNA replication occurs at the appropriate time and is accurately completed.
Are there any factors that can affect eukaryotic DNA replication?
Yes, various factors can influence eukaryotic DNA replication. Environmental factors, DNA damage, and mutations in replication-related genes can all impact the efficiency and fidelity of DNA replication.
Can errors occur during eukaryotic DNA replication?
Although eukaryotic DNA replication is highly accurate, errors can still occur. These errors can result in mutations that may have consequences for cellular function and potentially lead to genetic disorders or diseases.
How is eukaryotic DNA replication related to cancer?
Defects in DNA replication and repair mechanisms can contribute to the development of cancer. Mutations in genes involved in replication and checkpoint control can disrupt the normal regulation of cell growth, leading to uncontrolled cell division and tumor formation.
References
- Bhagavan, N. V., & Ha, C.-E. (2015). DNA Replication, Repair, and Mutagenesis. Essentials of Medical Biochemistry, 401–417. doi:10.1016/b978-0-12-416687-5.00022-1
- Priego, Sara & Gambus, Aga. (2015). Regulation of Unperturbed DNA Replication by Ubiquitylation. Genes. 6. 451-68. 10.3390/genes6030451.
- Riera et al. (2017) From structure to mechanism- understanding initiation of DNA replication. Genes Dev. 31(11): 1073-1088. doi: 10.1101/gad.298232.117
- Bhagavan & Ha (2015) Chapter 22 – DNA Replication, Repair, and Mutagenesis. Essentials of Medical Biochemistry (Second Edition) With Clinical Cases. 401-417. https://doi.org/10.1016/B978-0-12-416687-5.00022-1
- Tanaka et al. (2011) Sld7, an Sld3-associated protein required for efficient chromosomal DNA replication in budding yeast. EMBO J. 30(10): 2019-30. doi: 10.1038/emboj.2011.115. Epub 2011 Apr 12.
- Kamimura et al. (1998) Sld2, Which Interacts with Dpb11 in Saccharomyces cerevisiae, Is Required for Chromosomal DNA Replication. Mol Cell Biol. 1998 Oct; 18(10): 6102–6109.
- Georgescu et al. (2017) Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. PNAS 114 (5): E697-E706; first published January 17, 2017 https://doi.org/10.1073/pnas.1620500114.
- Watase et al. (2012) Mcm10 Plays a Role in Functioning of the Eukaryotic Replicative DNA Helicase, Cdc45-Mcm-GINS. Current Biology 22(4) 343-349.
- Bambara et al. (1997) Enzymes and Reactions at the Eukaryotic DNA Replication Fork. J Biol Chem. 272(8):4647-50.
- Abmayr and Workman (2012) Holding on through DNA Replication: Histone Modification or Modifier? Cell. 150(5):875-7.
- Verhoeven et al (2014). Cellular aging in depression: Permanent imprint or reversible process?: An overview of the current evidence, mechanistic pathways, and targets for interventions. BioEssays 36(10):968-78.
- Bailey et al (2015) Termination of DNA replication forks: “Breaking up is hard to do”.Nucleus 6 (3):187-196.
- Dewar & Walter (2017) Mechanisms of DNA replication termination. Nature Reviews Molecular Cell Biology 18:507–516.
- Aulinas (2013) Telomeres, aging and Cushing’s syndrome: Are they related? Endocrinology and Nutrition (English Edition) 60(6): 329-335.
- Boehm et al. (2016) The Many Roles of PCNA in Eukaryotic DNA Replication. Enzymes 39:231-54.
- https://en.wikipedia.org/wiki/Origin_recognition_complex
- https://biocyclopedia.com/index/genetics/chemistry_of_the_gene_synthesis_modification_and_repair_of_dna/different_steps_involved_in_eukaryotic_dna_replication.php
- https://www.thesciencenotes.com/dna-replication-in-eukaryotes/
- https://thebiotechnotes.com/2019/07/26/dna-replication-in-eukaryotes-elongation-and-termination/
- https://pressbooks-dev.oer.hawaii.edu/biology/chapter/dna-replication-in-eukaryotes/
- https://openoregon.pressbooks.pub/mhccmajorsbio/chapter/dna-replication-in-eukaryotes/
- https://www.jove.com/science-education/10789/replication-in-eukaryotes