What is DNA polymerase III (holoenzyme)?
- DNA polymerase III holoenzyme, a pivotal entity in prokaryotic DNA replication, was first identified by researchers Thomas Kornberg and Malcolm Gefter in 1970. This enzyme complex boasts a remarkable processivity, signifying its ability to incorporate a vast number of nucleotides in a single binding event. Notably, during the intricate process of E.coli genome replication, DNA polymerase III holoenzyme collaborates seamlessly with four other distinct DNA polymerases: Pol I, Pol II, Pol IV, and Pol V.
- One of the salient features of the DNA Pol III holoenzyme is its intrinsic proofreading capability. This function is facilitated by its exonuclease activity, which operates in a 3’→5′ direction.
- Concurrently, the enzyme synthesizes DNA in the 5’→3′ direction. Such dual functionality ensures the fidelity of DNA replication by meticulously rectifying any errors that might occur during the replication process.
- Furthermore, DNA Pol III is an integral part of the replisome, a sophisticated molecular machinery situated at the replication fork.
- This strategic positioning underscores its central role in ensuring the accurate and efficient replication of the prokaryotic genome. In essence, the DNA polymerase III holoenzyme is not merely an enzyme but a cornerstone of prokaryotic DNA replication, safeguarding the genetic integrity of the organism.
Definition of DNA polymerase III (holoenzyme)
DNA polymerase III holoenzyme is the primary enzyme complex in prokaryotes responsible for DNA replication, possessing both synthesis and proofreading capabilities to ensure accurate replication of the genome.
Components of DNA polymerase III (holoenzyme)
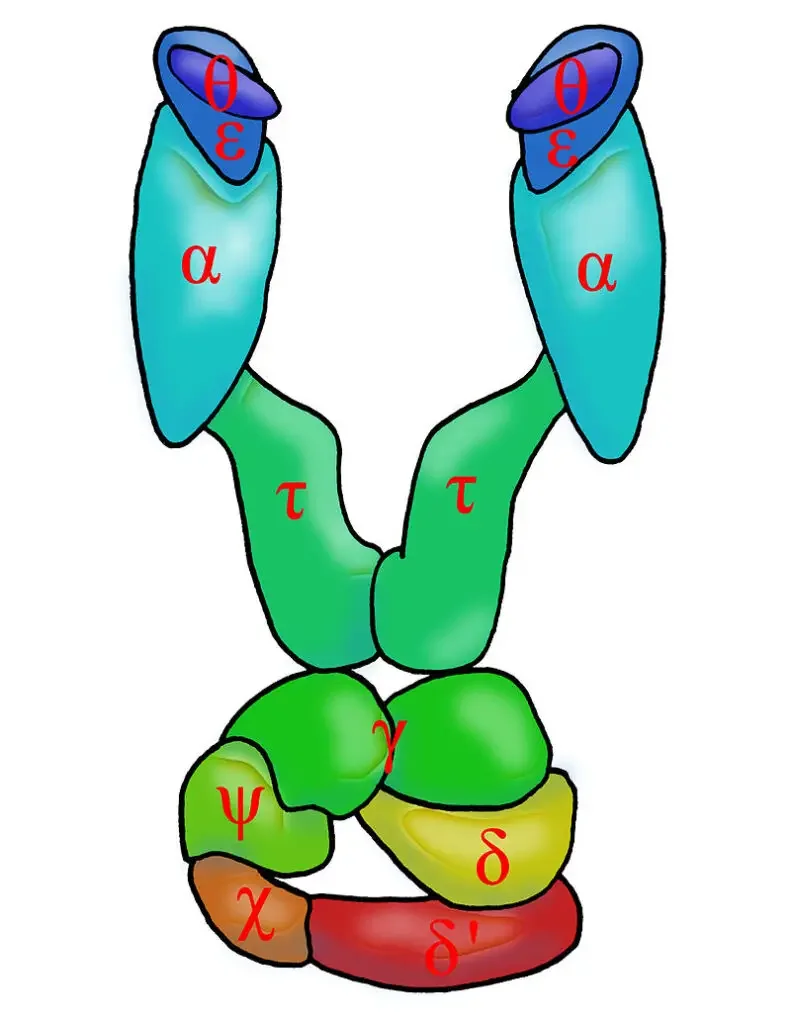
The DNA polymerase III holoenzyme, a central player in prokaryotic DNA replication, is a multifaceted complex composed of several distinct subunits, each with its specific function. Delving into its intricate architecture:
- Core Enzymes: The holoenzyme houses two DNA Pol III enzymes. Each of these enzymes is a triad of α, ε, and θ subunits.
- α subunit (dnaE gene product): This subunit is endowed with the polymerase activity, facilitating the synthesis of the DNA strand.
- ε subunit (encoded by dnaQ): This component possesses the 3’→5′ exonuclease activity, playing a pivotal role in proofreading and ensuring the fidelity of DNA replication.
- θ subunit (product of holE): Acting in synergy with the ε subunit, the θ subunit augments its proofreading capabilities.
- β units (encoded by dnaN): These are sliding DNA clamps. Their primary role is to anchor the polymerase to the DNA, ensuring its steadfastness during replication.
- τ units (product of dnaX): These units serve as bridges, dimerizing two core enzymes, thereby facilitating their coordinated function.
- γ unit (also a product of dnaX): This unit functions as a clamp loader, specifically for the lagging strand’s Okazaki fragments. It aids the β subunits in assembling and securing their position on the DNA. Intriguingly, the γ unit itself is a composite of five subunits: three γ, one δ (holA), and one δ’ (holB). The δ subunit has a specialized role in the replication of the lagging strand.
- Χ (encoded by holC) and Ψ (product of holD): These two components form a 1:1 complex and have an affinity for either γ or τ. Notably, the Χ subunit can orchestrate the transition from RNA primer synthesis to DNA synthesis.
In summation, the DNA polymerase III holoenzyme is a sophisticated ensemble of subunits, each contributing to the enzyme’s overarching function of accurate and efficient DNA replication.
Subunits and Subassembly of Pol III Holoenzyme
The Pol III Holoenzyme (Pol III HE) is a sophisticated molecular machinery that plays a pivotal role in DNA replication, particularly in the bacterium E. coli. Its intricate functionality is attributed to its subunit composition and the manner in which these subunits assemble.
- Subunit Composition and Assembly: The Pol III HE in E. coli is a conglomerate of ten distinct polypeptides, culminating in a 17-subunit complex. This assembly remains robust during its participation in the replisome at the replication fork. However, during purification, Pol III HE can disintegrate into free subunits and various subassemblies. These subassemblies have been instrumental in elucidating the biochemical roles of each subunit and the overall architecture of the holoenzyme.
- Pol III Core: Central to the holoenzyme is the α subunit, which embodies the DNA polymerase activity. This subunit, in conjunction with one molecule each of the ε and τ subunits, forms the Pol III core. The α subunit showcases a quintessential structure, reminiscent of a right hand, comprising palm, thumb, and finger domains. This structure is ubiquitous among DNA polymerases. The palm domain houses the polymerase motif, a sequence crucial for the active center of DNA polymerases. While the basic structure of DNA polymerases is conserved, they are categorized into six families (A, B, C, D, X, and Y) based on structural similarities. The α subunit of Pol III is a member of the family C, which is more akin to family X than family B. Unlike family B DNA polymerases, the α subunit of Pol III lacks the 3’→5′ exonuclease activity essential for proofreading. The ε subunit in the Pol III core serves as the editing exonuclease, ensuring accurate DNA synthesis.
- τ Subunit and Pol III’ (prime): The τ subunit, the second-largest in the ensemble, forms a stable homodimer. It plays a dual role: facilitating the dimerization of the Pol III core and serving as a bridge between dimeric polymerases and other auxiliary subunits. The resulting assembly, termed Pol III’, comprises two Pol III cores and two τ subunits.
- β Subunit and Pol III: The β subunit, known for its propensity to dissociate from the holoenzyme, forms a homodimer with a unique crescent shape. This dimer encircles the DNA and slides along it, tethering the α subunit to the template DNA. The β subunit’s interaction with other proteins and its sliding-clamp nature underpin the high processivity of Pol III HE.
- DnaX Complex: The DnaX complex, pivotal for loading the β clamp onto DNA, consists of the τ, γ, δ, and δ’ subunits. The τ and γ subunits, both products of the dnaX gene, are part of the AAA+ ATPase family. ATP binding and hydrolysis are central to the clamp-loading action of this complex.
- Pol III HE in Other Bacteria: While the architecture of Pol III HE in E. coli is well-understood, genes encoding its subunits have been identified in various eubacterial genomes. Although there’s divergence in some subunits across species, the fundamental architecture and functions of Pol III HE remain conserved across eubacteria.
In summation, the Pol III Holoenzyme is a marvel of molecular biology, with its subunits and subassemblies working in concert to ensure accurate and efficient DNA replication.
Activity of DNA polymerase III (holoenzyme)
DNA polymerase III holoenzyme, a central catalyst in prokaryotic DNA replication, exhibits remarkable enzymatic kinetics, synthesizing nucleotide sequences at an impressive rate of approximately 1000 nucleotides per second. The activity of this enzyme is initiated post the strand separation at the replication origin. However, it’s imperative to note that DNA synthesis cannot commence de novo. Instead, an RNA primer, synthesized by primase, provides a complementary sequence to a segment of the single-stranded DNA, setting the stage for DNA synthesis.
Illustratively:
Direction of Synthesis: -------->
* * * *
RNA Sequence: ! ! ! ! _ _ _ _
_ _ _ _ | RNA | <-- Ribose-phosphate backbone of RNA
Nucleotide Sequence: G U A U | Pol | <-- RNA primer
* * * * |_ _ _ _| <-- Hydrogen bonds
Template ssDNA: C A T A G C A T C C <-- Template single-stranded DNA
_ _ _ _ _ _ _ _ _ _ <-- Deoxyribose-phosphate backbone of DNA
As the replication machinery advances, DNA polymerase III encounters the RNA primer and commences the DNA synthesis, extending from the 3’OH end of the primer. This synthesis can manifest as either a continuous or discontinuous strand, contingent on whether it transpires on the leading or lagging strand, the latter being characterized by Okazaki fragments. The enzyme’s high processivity, a testament to its efficiency, is attributed in part to the β-clamps, which firmly grasp the DNA strands, ensuring steadfast synthesis.
Direction of Synthesis: ----------->
* * * *
RNA-DNA Hybrid: ! ! ! ! $ $ $ $ $ $ _ _ _ _
_ _ _ _ _ _ _ _ _ _| DNA | <-- Deoxyribose-phosphate backbone of DNA
Nucleotide Sequence: G U A U C G T A G G| Pol | <-- RNA primer transitioning to DNA
* * * * * * * * * *|_III_ _| <-- Hydrogen bonds
Template ssDNA: C A T A G C A T C C <-- Template single-stranded DNA
_ _ _ _ _ _ _ _ _ _ <-- Deoxyribose-phosphate backbone of DNA
Subsequent to the replication of the intended DNA segment, the RNA primer is excised by DNA polymerase I through nick translation. This pivotal step facilitates DNA ligase to bridge the gap between the nascent fragment and the preceding DNA strand, ensuring continuity. The concerted actions of DNA polymerases I & III, in synergy with a plethora of other enzymes, underscore the precision and efficiency inherent to DNA replication.
Functions of DNA polymerase III (holoenzyme)
DNA polymerase III holoenzyme (Pol III HE) plays a central role in bacterial DNA replication, particularly in Escherichia coli (E. coli). Its functions are multifaceted and crucial for the accurate and rapid synthesis of DNA. Here are the primary functions of DNA polymerase III holoenzyme:
- DNA Synthesis: The primary function of Pol III HE is the elongation of the DNA chain during replication. It adds nucleotides to the growing DNA strand in a 5’→3′ direction, ensuring the accurate duplication of the bacterial genome.
- High Processivity: Pol III HE can add thousands of nucleotides to the growing DNA strand without dissociating, thanks to its high processivity. This feature is largely attributed to the β-clamp, which holds the polymerase onto the DNA.
- Proofreading: Pol III HE has an intrinsic 3’→5′ exonuclease activity, which allows it to proofread each nucleotide it adds. If an incorrect nucleotide is incorporated, the enzyme can remove it, ensuring a high fidelity of DNA replication.
- Interaction with the Replisome: Pol III HE is a component of the larger replisome complex, which is responsible for coordinated DNA replication at the replication fork. It works in tandem with other enzymes and proteins to ensure smooth and synchronized DNA synthesis of both the leading and lagging strands.
- RNA Primer Recognition: DNA synthesis cannot start de novo. An RNA primer, synthesized by primase, provides a starting point. Pol III HE recognizes this primer and begins DNA synthesis from this point.
- Coordination of Leading and Lagging Strand Synthesis: Pol III HE is involved in the simultaneous synthesis of both the leading strand (continuous synthesis) and the lagging strand (discontinuous synthesis, forming Okazaki fragments). The holoenzyme’s dimeric nature allows it to coordinate the synthesis of both strands efficiently.
- Interaction with Other Proteins: Pol III HE interacts with various other proteins involved in DNA replication, such as the τ and γ complexes, ensuring efficient DNA loading, stabilization, and synthesis.
In essence, DNA polymerase III holoenzyme is not just a mere enzyme but a sophisticated molecular machine that ensures the rapid, accurate, and efficient replication of the bacterial genome.
Quiz
What is the primary role of DNA polymerase III holoenzyme in bacteria?
a) RNA synthesis
b) DNA repair
c) DNA replication
d) Protein synthesis
[expand title=”Show answer” swaptitle=”Hide answer”] c) DNA replication [/expand]
Which subunit of DNA polymerase III holoenzyme is responsible for its DNA polymerase activity?
a) ε subunit
b) β subunit
c) τ subunit
d) α subunit
[expand title=”Show answer” swaptitle=”Hide answer”] d) α subunit [/expand]
Which direction does DNA polymerase III holoenzyme synthesize DNA?
a) 3’→5′
b) 5’→3′
c) 5’→5′
d) 3’→3′
[expand title=”Show answer” swaptitle=”Hide answer”] b) 5’→3′ [/expand]
Which subunit of DNA polymerase III holoenzyme has 3’→5′ exonuclease activity for proofreading?
a) α subunit
b) ε subunit
c) τ subunit
d) β subunit
[expand title=”Show answer” swaptitle=”Hide answer”] b) ε subunit [/expand]
The β-clamp of DNA polymerase III holoenzyme plays a crucial role in:
a) DNA synthesis
b) RNA primer recognition
c) High processivity
d) Interaction with the replisome
[expand title=”Show answer” swaptitle=”Hide answer”] c) High processivity [/expand]
Which enzyme synthesizes the RNA primer that DNA polymerase III holoenzyme recognizes to start DNA synthesis?
a) Helicase
b) Ligase
c) Primase
d) Topoisomerase
[expand title=”Show answer” swaptitle=”Hide answer”] c) Primase [/expand]
Which of the following is NOT a function of DNA polymerase III holoenzyme?
a) Proofreading
b) RNA synthesis
c) Interaction with the replisome
d) Coordination of leading and lagging strand synthesis
[expand title=”Show answer” swaptitle=”Hide answer”] b) RNA synthesis [/expand]
Which family does the α subunit of DNA polymerase III belong to?
a) Family A
b) Family B
c) Family C
d) Family X
[expand title=”Show answer” swaptitle=”Hide answer”] c) Family C [/expand]
The τ subunit of DNA polymerase III holoenzyme primarily facilitates:
a) DNA synthesis
b) Proofreading
c) Dimerization of the Pol III core
d) Interaction with the β-clamp
[expand title=”Show answer” swaptitle=”Hide answer”] c) Dimerization of the Pol III core [/expand]
Which subunit of DNA polymerase III holoenzyme encircles the DNA and slides along it during replication?
a) α subunit
b) ε subunit
c) β subunit
d) τ subunit
[expand title=”Show answer” swaptitle=”Hide answer”] c) β subunit [/expand]
FAQ
What is DNA polymerase III holoenzyme?
DNA polymerase III holoenzyme is the primary enzyme complex responsible for DNA replication in prokaryotes, particularly in E. coli.
How does DNA polymerase III holoenzyme differ from other DNA polymerases?
DNA polymerase III holoenzyme is unique in its structure, subunit composition, and high processivity. It is specifically designed for rapid and accurate replication of the bacterial genome.
What is the primary function of DNA polymerase III holoenzyme?
The primary function of DNA polymerase III holoenzyme is to synthesize DNA during replication, ensuring the accurate duplication of the bacterial genome.
Which subunit of DNA polymerase III holoenzyme is responsible for its DNA polymerase activity?
The α subunit is responsible for the DNA polymerase activity of the holoenzyme.
How does DNA polymerase III holoenzyme ensure the accuracy of DNA replication?
DNA polymerase III holoenzyme has an intrinsic 3’→5′ exonuclease activity, allowing it to proofread each nucleotide it adds. If an incorrect nucleotide is incorporated, the enzyme can remove it, ensuring high fidelity.
What role does the β-clamp play in the function of DNA polymerase III holoenzyme?
The β-clamp plays a crucial role in the high processivity of the enzyme. It holds the polymerase onto the DNA, allowing it to add thousands of nucleotides without dissociating.
Can DNA polymerase III holoenzyme start DNA synthesis on its own?
No, DNA polymerase III holoenzyme cannot start DNA synthesis de novo. It requires an RNA primer, synthesized by primase, to initiate DNA synthesis.
What is the significance of the τ subunit in DNA polymerase III holoenzyme?
The τ subunit facilitates the dimerization of the Pol III core, allowing the holoenzyme to coordinate the synthesis of both the leading and lagging strands simultaneously.
How does DNA polymerase III holoenzyme differ across bacterial species?
While the basic architecture and functions of Pol III HE are conserved across eubacteria, there can be variations in the subunit composition and specific functionalities in different bacterial species.
Why is DNA polymerase III holoenzyme considered a molecular machine?
DNA polymerase III holoenzyme is a complex ensemble of multiple subunits that work in concert to ensure rapid, accurate, and efficient replication of the bacterial genome, making it akin to a sophisticated molecular machine.
References
- Maki, H., & Furukohri, A. (2013). DNA Polymerase III, Bacterial. Encyclopedia of Biological Chemistry, 92–95. doi:10.1016/b978-0-12-378630-2.00310-8