What is DNA Library (Genomic, cDNA)?
- A DNA library is a collection of DNA fragments stored in vectors and kept inside host organisms. These fragments can either represent an entire genome or just the genes active at a specific time.
- Genomes are vast and complex. To make sense of them, breaking them down into smaller pieces helps. DNA libraries do just that, making the genome more manageable. They’re crucial for identifying, isolating, and studying specific genes.
- The birth of DNA libraries was driven by advances in molecular biology. Recombinant DNA technology, cloning vectors, and bacterial transformation techniques made it all possible.
- There are two main types of DNA libraries: genomic and cDNA.
- A genomic library contains DNA fragments that together represent the entire genome of an organism. The process starts with extracting DNA and cutting it into fragments using restriction enzymes. These fragments are then inserted into vectors with the help of DNA ligase. The vectors are introduced into host cells, often E. coli or yeast, which then carry and replicate the DNA fragments.
- The choice of vectors depends on the size of the genome being studied. Larger genomes need vectors that can hold larger inserts to reduce the number of vectors required for a complete library.
- Genomic libraries are key for sequencing projects. They were essential in sequencing the human genome and other model organisms. The first complete DNA-based genome sequence was achieved by Frederick Sanger in 1977, using a library of the bacteriophage phi X 174. This milestone paved the way for genome-wide studies and gene therapy research.
- A cDNA library, on the other hand, is made from reverse-transcribed mRNA, focusing only on expressed genes. These libraries lack non-coding regions like introns. They are typically stored in plasmid vectors and contain smaller fragments than genomic libraries.
- Both types of DNA libraries are fundamental tools in genetic research, helping scientists explore gene functions, discover new genes, and understand genetic diseases.
Types of DNA Library
There are two main types of DNA libraries: genomic libraries, and cDNA libraries.
1. Genomic Library
- A genomic library is a collection of DNA fragments that cover an organism’s entire genome.
- This includes both the coding regions (genes) and the noncoding regions (regulatory elements, introns).
- Genomic libraries are used for genome mapping and comparative genomics.
- They help study regulatory elements and noncoding sequences involved in gene expression.
- Creating and maintaining genomic libraries can be complex and resource-intensive.
- Handling large DNA fragments is challenging.
2. cDNA Library
- A cDNA library is a collection of cDNA molecules made from mRNA.
- Unlike genomic libraries, cDNA libraries only represent the expressed genes of an organism.
- Non-expressed genomic regions like introns and other noncoding sequences are excluded.
- cDNA libraries are great for studying gene expression, protein functions, and making recombinant proteins.
- Since they exclude noncoding regions, they offer a more focused view of expressed genes.
- However, cDNA libraries can’t study gene regulation well due to the lack of regulatory sequences.
- They also have limited gene diversity and are biased towards highly expressed genes.
- cDNA libraries are usually made from eukaryotes to study expressed genes.
- Prokaryotes don’t need cDNA libraries because their genomic DNA directly corresponds to their mRNA.
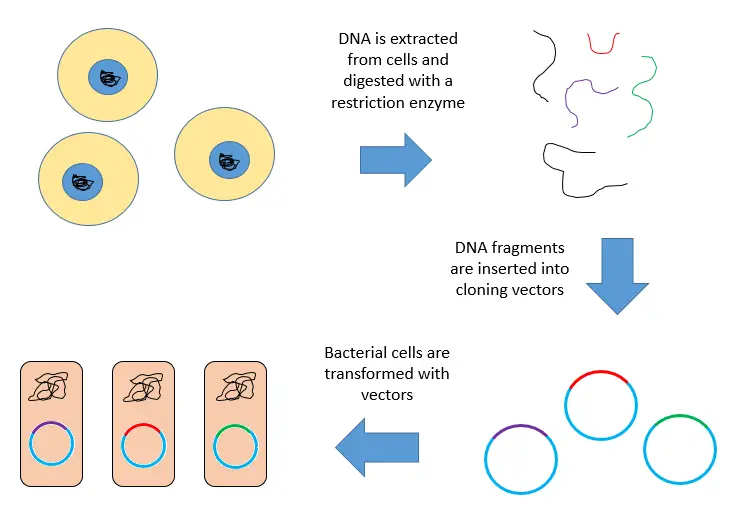
Genomic Library Construction Steps
- Extract and purify DNA: Start by getting the genomic DNA out of the organism and purifying it.
- Digest with restriction enzyme: Cut the DNA into fragments using a restriction enzyme. These fragments are similar in size and contain one or more genes.
- Insert into vectors: Put these DNA fragments into vectors that have also been cut with the same restriction enzyme.
- Seal with DNA ligase: Use DNA ligase to seal the DNA fragments into the vector. This creates a large pool of recombinant molecules.
- Transformation: Introduce these recombinant molecules into a host bacterium through transformation.
- Create the library: This process results in a genomic library, where each bacterium contains a different recombinant molecule representing a fragment of the organism’s genomic DNA.
Genomic Library Construction Steps
Creating a cDNA library begins with mRNA, which carries genetic instructions from DNA to ribosomes for protein synthesis. Here’s how it’s done:
- Isolation of mRNA
- First, isolate total mRNA from a specific cell type or tissue of interest.
- To enhance the library quality, remove abundant tRNAs and rRNAs using specialized kits.
- Use an oligo-dT column to purify mRNA, which binds specifically to the poly A tail of mRNA molecules.
- Techniques like density gradient centrifugation can also increase mRNA yield.
- Synthesis of the First Strand of cDNA
- Use a short oligo (dT) primer to bind to the poly A tail of mRNA.
- Reverse transcriptase then synthesizes the first strand of cDNA using mRNA as a template.
- Remove the mRNA from the cDNAhybrid to obtain a single-stranded cDNA molecule.
- Generation of the Second Strand of cDNA
- Convert the single-stranded cDNA into double-stranded cDNA using reverse transcriptase or E. coli DNA polymerase.
- Minimize amplification cycles to avoid bias in the library.
- Incorporation of cDNA into a Vector
- Trim the double-stranded cDNA to obtain blunt-ended molecules, then add C residues using terminal transferase.
- Ligase short restriction-site linkers to both ends for efficient ligation into a vector.
- Cloning of cDNAs
- Use phage insertion vectors for cloning cDNAs.
- Phage vectors are advantageous for cloning low-abundance mRNAs due to their ability to produce recombinant phages through in vitro packaging.
- They also allow efficient handling and storage of a large number of clones compared to bacterial colonies carrying plasmids.
Differences between Genomic Library and cDNA Library
Here’s a straightforward breakdown of the disparities between genomic libraries and cDNA libraries:
cDNA Library
- What it is: Collection of clones containing complementary DNA to mRNA of an organism.
- What it contains: Represents genes expressed in a specific cell at a particular time.
- Size: Smaller compared to genomic DNA library.
- Coding and non-coding sequences: Contains sequences observed on mRNA only, not the entire gene.
- Important substances required: Uses reverse transcriptase enzyme.
- Number of recombinants to be screened: Fewer compared to genomic DNA library.
- Material required to start: mRNA.
- Expression in prokaryotic system: Can be directly expressed as it has only coding sequences.
- Role in reverse transcriptase: Involved in the synthesis of the first cDNA strand.
- Vectors used: Phagemids, Plasmids, Lambda phage for harboring smaller fragments due to the absence of introns.
Genomic DNA Library
- What it is: Collection of clones containing the complete genomic DNA of an organism.
- What it contains: Represents all genes, including coding and non-coding sequences like introns and regulatory elements.
- Size of DNA fragments: Vast in comparison to cDNA library, ranging from thousands to millions of base pairs.
- Important substances required: Uses ligases and restriction endonucleases.
- Number of recombinants to be screened: Greater compared to cDNA library.
- Material required to start: DNA.
- Expression in prokaryotic system: Challenging due to the presence of introns.
- Role in reverse transcriptase: Not involved.
- Vectors used: Cosmids, plasmids, Lambda phage, YAC, BAC to harbor larger fragments.
Characteristics
- Genomic Library: Contains DNA fragments representing the entire genome, including coding and non-coding regions.
- cDNA Library: Consists only of cDNA sequences that are actively expressed.
- Size of DNA fragments: Genomic libraries have larger DNA fragments, whereas cDNA libraries contain smaller fragments.
- Source of DNA: Genomic libraries use total genomic DNA extracted from cells or tissues, while cDNA libraries use mRNA converted to cDNA.
- Vectors used: Genomic libraries use vectors capable of holding large DNA fragments, while cDNA libraries typically use plasmid vectors suitable for smaller cDNA fragments.
- Application: Genomic libraries are used for genomic mapping, comparative genomics, and studying gene regulation. cDNA libraries are particularly useful for studying gene expression patterns and the functions of expressed genes.
| Characteristic | Genomic Library | cDNA Library |
|---|---|---|
| What it is | Collection of clones containing the complete genomic DNA of an organism | Collection of clones containing complementary DNA to mRNA |
| Content | Represents all genes, including coding and non-coding sequences | Represents genes expressed in a specific cell at a particular time |
| Size of DNA fragments | Larger, ranging from thousands to millions of base pairs | Smaller compared to genomic DNA library |
| Coding and non-coding regions | Contains both coding and non-coding regions (introns, regulatory elements) | Contains sequences observed on mRNA only, not the entire gene |
| Source of DNA | Total genomic DNA extracted from cells or tissues | mRNA converted to cDNA |
| Vectors used | Cosmids, plasmids, Lambda phage, YAC, BAC for larger DNA fragments | Phagemids, Plasmids, Lambda phage for smaller cDNA fragments |
| Important substances required | Ligases and restriction endonucleases | Reverse transcriptase enzyme |
| Number of recombinants to be screened | Greater compared to cDNA library | Fewer compared to genomic DNA library |
| Material required to start | DNA | mRNA |
| Expression in prokaryotic system | Challenging due to the presence of introns | Can be directly expressed as it has only coding sequences |
| Role in reverse transcriptase | Not involved | Involved in the synthesis of the first cDNA strand |
| Application | Genomic mapping, comparative genomics, studying gene regulation | Studying gene expression patterns and functions of expressed genes |
Applications of DNA Library
Applications of Genomic Libraries:
- Physical Maps of Genomes: Essential for constructing maps that detail the layout and structure of genes within an organism’s genome.
- Study of Non-Coding Regions: Useful for investigating non-coding regions, regulatory elements, and sequences that may not be actively transcribed.
- Comparative Genomics: Facilitates comparison of genomic libraries from different species to study evolutionary relationships and genetic variations.
- Identification of Disease Genes: Helps in identifying genetic mutations and genes associated with diseases, aiding in understanding the genetic basis of various disorders.
Applications of cDNA Libraries:
- Study of Expressed Genes: Useful for studying actively expressed genes in different tissues under specific conditions, allowing for identification and cloning of these genes.
- Comparative Gene Expression: Enables comparison of gene expression profiles between different species, contributing to the study of evolutionary biology.
- Production of Recombinant Proteins: Used in biotechnology to produce proteins of interest for various applications.
- Disease-Related Research: Helps identify and study genes associated with diseases, aiding in the development of diagnostic markers and potential therapeutic targets.
References
- Wikipedia contributors. (2024, May 25). Genomic library. In Wikipedia, The Free Encyclopedia. Retrieved 06:55, July 4, 2024, from https://en.wikipedia.org/w/index.php?title=Genomic_library&oldid=1225627387
- Chang, Liu., Wang, Xuan., Jiu‑Bo, Tian., Pei‑Yuan, Ma., Fan‑Xiu, Meng., Qi, Zhang., Bao‑Feng, Yu., Rui, Guo., Zhi‑Zhen, Liu., Hai-Long, Wang., Jun, Xie., Niu‑Liang, Cheng., Jian‑Hua, Wang., Bo, Niu., Gong‑Qin, Sun. (2019). Construction of a cDNA library and preliminary analysis of the expressed sequence tags of the earthworm Eisenia fetida (Savigny, 1826).. Molecular Medicine Reports, doi: 10.3892/MMR.2019.9938
- Maarten, Kooiker., Gang-Ping, Xue. (2014). cDNA library preparation.. Methods of Molecular Biology, doi: 10.1007/978-1-62703-715-0_5
- Naiara, Rodríguez-Ezpeleta., Shona, Teijeiro., Lise, Forget., Gertraud, Burger., B., Franz, Lang. (2009). Construction of cDNA Libraries: Focus on Protists and Fungi. Methods of Molecular Biology, doi: 10.1007/978-1-60327-136-3_3
- Shao-Yao, Ying. (2004). Complementary DNA libraries: an overview.. Molecular Biotechnology, doi: 10.1385/MB:27:3:245
- Dario, Neri. (2017). Twenty-five Years of DNA-Encoded Chemical Libraries.. ChemBioChem, doi: 10.1002/CBIC.201700130
- Julie, M., Struble., P., Handke., Ryan, T., Gill. (2009). Genome Sequence Databases: Genomic, Construction of Libraries. doi: 10.1016/B978-012373944-5.00026-2
- Jennifer, Gebetsberger., Roger, Fricker., Norbert, Polacek. (2015). cDNA library generation for the analysis of small RNAs by high-throughput sequencing.. Methods of Molecular Biology, doi: 10.1007/978-1-4939-2547-6_13
- Michael, P., Starkey., Yaghesh, Umrania., Christopher, R., Mundy., Martin, J., Bishop. (1998). Reference cDNA library facilities available from European sources. Molecular Biotechnology, doi: 10.1007/BF02752696
- https://www.thermofisher.com/in/en/home/life-science/cloning/cloning-applications/library-construction.html
- https://byjus.com/neet/difference-between-cdna-and-genomic-dna-library/
- https://www.creative-biogene.com/support/cdna-library-construction-protocol.html