What is Transcription?
- Transcription is the process by which the genetic information stored on DNA strands is transferred into an RNA-RNA strand via the polymerization process caused by enzymes known as DNA-dependent polymerases.
- This is the very first stage of gene expression in which the information is transferred between structures to the next.
- In the process of transcription, molecules of RNA get activated, stretched and then terminated. The RNA produced is non-genetic RNA.
- Transcription is an essential step for translation. It happens in the event of a requirement for a specific gene at a particular time to the specific tissue.
- One strand of DNA known as the template strand, gets replicated during transcription. that resulting RNA strands form mono-stranded messenger-RNA (mRNA).
- The transcription of a particular gene occurs close to the chromosomal location of the gene. This is generally a narrow section of the chromosome.
- The entire process of transcription is highly controlled which is controlled and catalyzed by enzyme DNA-dependent RNA polymerase.
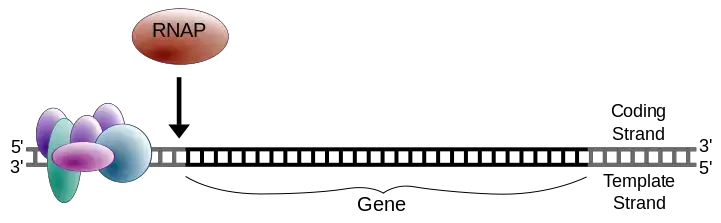
- The first step is the identification of DNA sequences, also known as promoter sequences, which signify the start for the genetic code.
- The process is followed by the division of two DNA strands and the replication of one the DNA strands using the polymerase RNA.
- The RNA polymerase that is found in Eukaryotes is different and more complicated as compared to prokaryotes.
- The sequence that is created after replication is a complement to that of the original template since it follows the complementary bases pairing guidelines of DNA with the exception that thymine is replaced by uracil.
- In prokaryotes, the entire process is controlled by proteins that act as signaling or operators. They end it by blocking RNA polymerase after the process is completed.
- In eukaryotes, the various proteins referred to as transcription factors play a role in the control of transcription.
- Additionally, the post-transcriptional alteration is also seen in eukaryotes in which the pre-mRNA (the product that transcription) is altered through the process of splicing, which occurs before the mature mRNA gets to the ribosomes that are used for translation.
- The mRNA produced functions as a guideline for protein production in transcription.
- The sequence of DNA selected to be used for transcription, rRNA as well as tRNA synthesis can also take place.
- The transcription process takes place inside the nucleus of the eukaryotes as well as in the in the cytoplasm of prokaryotes where the enzymes as well as transcriptional factors are present.
- It is blocked by certain antibiotics such as rifampicin and 8Hydroxyquinoline.
- The process is detected with methods such as DNA microarrays, RTPCR hybridization in situ, and northern Blotting.
Stages of Transcription
- Pre-Initiation:
- In eukaryotes, DNA is stored in the nucleus and is tightly packed in chromatin.
- The RNA polymerase σ subunit binds to the promoter region at the 5′ end of the DNA strand.
- The DNA strands separate, exposing the template strand for transcription.
- Promoter sequences, such as TATAAT and TTGACA in prokaryotes, and TATAAAA and GGCCAATCT in eukaryotes, are important for successful initiation.
- In eukaryotes, additional transcription factors aid in the binding of RNA polymerase to the promoter region.
- Initiation:
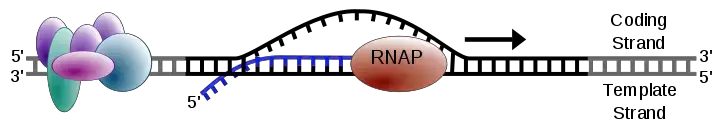
- RNA polymerase catalyzes the initiation of transcription.
- The first complementary 5′-ribonucleoside triphosphate is introduced, and subsequent complementary ribonucleotides are added in a 5′ to 3′ direction, joined by phosphodiester bonds.
- The RNA nucleotide bases are complementary to the DNA bases, except that RNA uses uracil (U) instead of thymine (T).
- Elongation:
- The σ subunit dissociates from the DNA strand, allowing the growing RNA strand to separate from the DNA template.
- Chain elongation occurs as the core enzyme facilitates the addition of complementary ribonucleotides, extending the RNA strand in a 5′ to 3′ direction.
- Termination:
- Termination occurs when the core enzyme encounters a termination sequence.
- In prokaryotes, a termination factor called rho (ρ) can assist in termination.
- The RNA transcript forms a hairpin secondary structure, folding back on itself with hydrogen bonds.
- In eukaryotes, termination involves an additional step called polyadenylation, where a tail of multiple adenosine monophosphates is added to the RNA strand.
It’s important to note that the provided content mainly focuses on the process of transcription in prokaryotes and eukaryotes, and does not cover the subsequent processing and modifications of the RNA transcript.
What is Reverse Transcription?
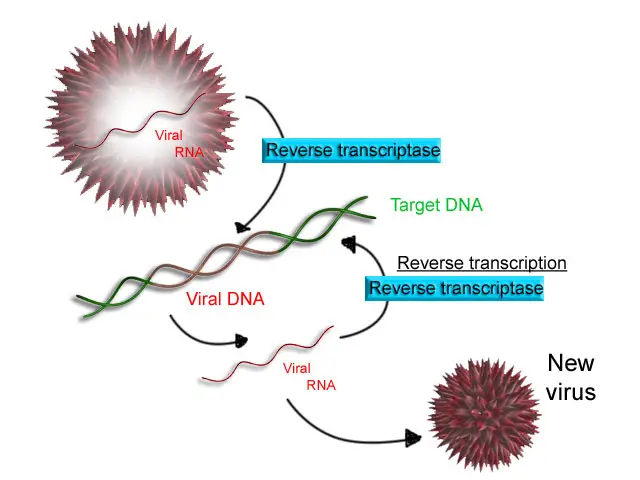
Reverse transcription is a process by which RNA molecules are reverse transcribed into complementary DNA (cDNA). It is catalyzed by an enzyme called reverse transcriptase. Reverse transcription is a characteristic feature of retroviruses, which use reverse transcriptase to convert their RNA genome into DNA, allowing integration of the viral genetic material into the host cell’s genome.
The process of reverse transcription involves the following steps:
- Initiation: Reverse transcriptase binds to a specific region of the RNA molecule called the primer binding site. It synthesizes a short stretch of complementary DNA known as the primer using ribonucleotides as a template.
- Reverse Transcription: The primer serves as a starting point for reverse transcriptase to synthesize the complementary DNA strand. The enzyme moves along the RNA molecule, synthesizing a complementary DNA strand using deoxyribonucleotides (dNTPs) and the RNA template as a guide.
- RNA Degradation: Once the reverse transcription is complete, the RNA portion of the RNA-DNA hybrid is degraded by the enzyme’s ribonuclease H (RNase H) activity. RNase H cleaves the RNA strand, leaving behind the newly synthesized cDNA strand.
- Second Strand Synthesis: The single-stranded cDNA serves as a template for the synthesis of the complementary DNA strand. Reverse transcriptase possesses DNA polymerase activity, which catalyzes the synthesis of the second DNA strand using the initial cDNA strand as a template.
The resulting product of reverse transcription is a double-stranded cDNA molecule, which can be further utilized for various applications in molecular biology, such as cloning, gene expression analysis, and genotyping. Reverse transcription is particularly useful in studying RNA viruses, analyzing gene expression patterns, and performing techniques like quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
What is Translation?
- The process of translation is of protein synthesis, where the RNA information is expressed as polypeptide chains.
- It is the second and final step in the process of expression of genes where the information encoded by the mRNA sequence is converted into the amino acid sequence.
- The beginning point for translation lies in the messenger RNA that has formed through the process of transcription of a specific DNA sequence. Translation follows transcription.
- Although the information contained in the mRNA is used to create amino acid sequences other types of RNA, such as tRNA are also needed for the procedure.
- Similar to transcription, translation can also be controlled by a variety of elements and enzymes. The most crucial enzyme is aminoacetyl synthetase for tRNA.
- The process begins with the initial phase, which involves binding of mRNA to Ribosomes. Then comes the transfer and attachment of activated amino acid on the transcriptase.
- The next process is elongation. the two amino acids get linked by the peptide bond, as molecules mRNA and ribosomes shift in relation each other to enable the simultaneous translation of codons.
- After all codons have been translated, the resulting polypeptide sequence is separated from the complex of translation and ribosomes are released to start a new phase of translation.
- The termination is followed by post-translational modification where the polypeptide must be folded in order to get the three-dimensional shape. This happens in the endoplasmic-reticulum and Golgi apparatus in the cell. Consequently the polypeptide chains are transferred to these organelles.
- Some modifications may be chemical and require the attaching functional group to sequence of peptides.
- The process of translating is controlled by the binding of ribosomal subunits with this translation component. Ribosomes are enzymes that regulate different actions.
- In eukaryotes it is located in the ribosomes that are associated with the endoplasmic-reticulum, whereas in prokaryotes it is located within the cytoplasm.
- The translation is blocked by antibiotics such as tetracycline streptomycin and chloramphenicol anisomycin, Cycloheximide, and so on.
- The procedure of translating can also be identified using methods like western blotting, immunoblotting enzyme assays Protein sequencing, and so on.
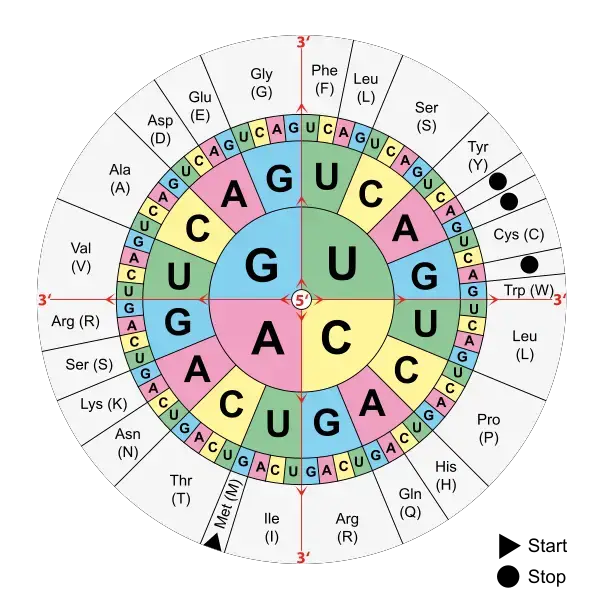
Stages of Translation
The stages of translation can be described as follows:
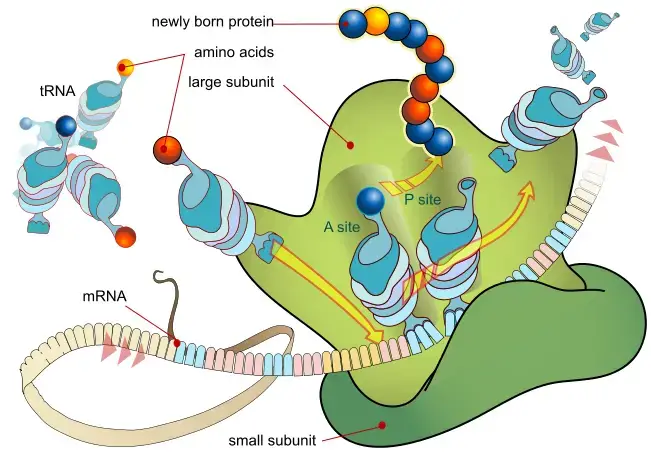
- Initiation:
- In both prokaryotes and eukaryotes, translation begins with the small ribosomal subunit binding to the 5′ end of the mRNA.
- Initiation factors facilitate the binding of the small ribosomal subunit to mRNA.
- A charged molecule of transfer RNA (tRNA) carrying methionine (in most cases) binds to the small ribosomal subunit.
- The large ribosomal subunit joins the complex, hydrolyzing GTP, and releasing initiation factors.
- Elongation:
- Elongation starts when both the small and large ribosomal subunits are bound to the mRNA.
- The tRNA carrying the amino acid that corresponds to the codon in the mRNA binds to the A site (aminoacyl site) of the ribosome.
- Peptidyl transferase, an enzyme within the ribosome, catalyzes the formation of a peptide bond between the amino acid on the tRNA in the A site and the growing polypeptide chain attached to the tRNA in the P site (peptidyl site).
- The tRNA in the P site is released, while the tRNA in the A site moves to the P site, and the A site is now available for the binding of the next tRNA.
- This process continues, with each new tRNA bringing its amino acid and forming a peptide bond with the growing polypeptide chain.
- Termination:
- Termination occurs when the ribosome encounters a stop codon on the mRNA.
- GTP-dependent release factors bind to the A site, causing the release of the completed polypeptide chain from the last tRNA.
- The ribosome complex dissociates, releasing the small and large ribosomal subunits from the mRNA.
- The polypeptide chain folds into its functional protein conformation.
It’s important to note that the process of translation involves the participation of various other molecules and factors, such as aminoacyl-tRNA synthetases, which link specific amino acids to their corresponding tRNA molecules, and GTP (guanosine-5′-triphosphate) for energy supply. Additionally, post-translational modifications may occur after translation to further modify and process the protein.
Differences between Transcription and Translation
- Localization:
- In prokaryotes, transcription and translation occur in the cytoplasm.
- In eukaryotes, transcription occurs in the nucleus, and translation occurs in ribosomes on the rough endoplasmic reticulum in the cytoplasm.
- Factors:
- Transcription: RNA polymerase and transcription factors are involved.
- Transcription can be inducible or constitutive.
- Translation: Ribosome, rRNA, and proteins are involved.
- Transcription: RNA polymerase and transcription factors are involved.
- Initiation:
- Transcription: RNA polymerase binds to the promoter region, forming a transcription initiation complex.
- Translation: The initiation complex is formed, including ribosome subunit, initiation factors, and methionine-carrying tRNA.
- Elongation:
- Transcription: RNA polymerase moves along the DNA template, producing a complementary RNA strand.
- Translation: Aminoacyl tRNA binds to the codon at the A-site, and a peptide bond is formed. The process proceeds in a 5′ to 3′ direction.
- Termination:
- Transcription: Prokaryotes can have Rho-independent or Rho-dependent termination. In eukaryotes, the RNA transcript is released and poly-adenylated.
- Translation: When a stop codon is encountered, the ribosome disassembles and releases the polypeptide.
- End Product:
- Transcription: RNA transcript types include mRNA, tRNA, rRNA, and non-coding RNA.
- Prokaryotes have polycistronic mRNA, while eukaryotes have monocistronic mRNA.
- Translation: The end product is a polypeptide chain, which undergoes folding and post-translational modifications to form a functional protein.
- Transcription: RNA transcript types include mRNA, tRNA, rRNA, and non-coding RNA.
- Post Process Modification:
- Eukaryotes undergo post-transcriptional modifications such as adding a 5′ cap, a 3′ poly tail, and splicing out introns.
- Prokaryotes lack this process.
- Post-translational modifications include phosphorylation, SUMOylation, disulfide bridge formation, farnesylation, etc.
- Antibiotics:
- Transcription inhibitors: Rifampicin (antibacterial), 8-Hydroxyquinoline (antifungal).
- Translation inhibitors: Anisomycin, cycloheximide, chloramphenicol, tetracycline, streptomycin, erythromycin, puromycin.
- Methods to measure and detect:
- Transcription:
- RT-PCR, DNA microarray, In-situ hybridization, Northern blot, RNA-Seq.
- Translation:
- Western blotting, immunoblotting, enzyme assay, protein sequencing, metabolic labeling, proteomics.
- Transcription:
- Crick’s central dogma:
- DNA → Transcription → RNA → Translation → Protein.
Key Differences between Transcription and Translation (Transcription vs Translation)
| Basis for Comparison | Transcription | Translation |
|---|---|---|
| Purpose | The purpose of transcription is to make RNA copies of individual genes that the cell can use in the biochemistry. | The purpose of translation is to synthesize proteins, which are used for millions of cellular functions. |
| Definition | Uses the genes as templates to produce several functional forms of RNA | Translation is the synthesis of a protein from an mRNA template. This is the second step of gene expression. Uses rRNA as assembly plant; and tRNA as the translator to produce a protein. |
| Gene expression | Transcription is the very first step in the process of expressing genes. | The translation process is the second and final stage of expression. |
| Occurs | Transcription occurs before translation. | The translation process begins after transcription. |
| Precursor | The transcription precursor is the antisense or non-coding DNA Strand. | The precursor to translation is the mRNA that results from transcription. |
| Material in raw form | The transcription raw material comprises the four bases of RNA: adenine, guanine, uracil, and cytosine. | The basic ingredients of translation are the twenty amino acids. |
| Initiation | Transcription is initiated by the recognition of specific DNA sequences, known as promoter sequences, which signals the start of gene activity. | Translation is initiated by the binding of mRNA to ribosomes. |
| Elongation | Elongation in transcription involves the addition of base pairs to the newly formed RNA sequence. | Elongation in translation occurs through the bonding of amino acids to the growing polypeptide chain. |
| Product | The end product of transcription is an mRNA-like molecule that is complementary to the DNA strand. | The result of translation is a peptide sequence encoded by the mRNA sequence. |
| Synthesis of | Transcription leads to the synthesis of RNA sequences. | Translation leads to protein synthesis. |
| Site | Transcription occurs in the nucleus of eukaryotes and in the cytoplasm of prokaryotes, where regulators and enzymes are present. | Translation occurs in the cytoplasm of prokaryotes and on the ribosomes lining the endoplasmic reticulum in eukaryotes. |
| Enzymes | The main enzymes involved in transcription are DNA-dependent polymerases. | The main enzyme responsible for translation is aminoacyl synthetase. |
| Regulation | Transcription is regulated by transcription factors in eukaryotes and through operons in prokaryotes. | Translation is regulated by the interaction of ribosomal units with the transcription complex. |
| Modifications | Post-transcriptional modifications involve processing of pre-mRNA, such as splicing, before mature mRNA is ready for translation. | Post-translational modifications require the stretching of polypeptide chains to create the three-dimensional structure of proteins. |
| Detection | Techniques such as DNA microarray, RT-PCR, in-situ hybridization, and northern blotting can be used to detect transcription. | Translation can be identified through methods such as western blotting, immunoblotting, enzyme assays, and protein sequencing. |
| Inhibition | Transcription can be blocked by certain antibiotics like rifampicin and 8-hydroxyquinoline. | Translation can be inhibited by antibiotics such as tetracycline, streptomycin, chloramphenicol, anisomycin, cycloheximide, etc. |
| Localization | Found in the nucleus of eukaryotes and in the cytoplasm of prokaryotes. | Found in the cytoplasm of prokaryotes and on ribosomes lining the endoplasmic reticulum in eukaryotes. |
| Purpose | The purpose of transcription is to make RNA copies of individual genes that the cell can use in biochemistry. | The purpose of translation is to synthesize proteins, which are used for millions of cellular functions. |
| Products | mRNA, tRNA, rRNA, and non-coding RNA (e.g., microRNA) | Proteins |
| Product processing | A 5’ cap is added, a 3’ poly A tail is added, and introns are spliced out. | A number of post-translational modifications occur, including phosphorylation, SUMOylation, disulfide bridges, and farnesylation. |
| Termination | RNA transcript is released and polymerase detaches from DNA. DNA rewinds itself into a double-helix and is unaltered throughout this process. | When the ribosome encounters one of the three stop codons, it disassembles the ribosome and releases the polypeptide. |
| Localization | Nucleus | Cytoplasm |
| Initiation | Occurs when RNA polymerase protein binds to the promoter in DNA and forms a transcription initiation complex. | Occurs when ribosome subunits, initiation factors, and tRNA bind to the mRNA near the AUG start codon. |
| Elongation | RNA polymerase elongates in the 5′ –> 3′ direction | The incoming aminoacyl t-RNA binds to the codon at the A-site, and a peptide bond is formed between the new amino acid and the growing chain. The peptide then moves one codon position to get ready for the next amino acid. It then proceeds in a 5′ to 3′ direction. |
| Antibiotics | Transcription is inhibited by rifampicin and 8-hydroxyquinoline. | Translation is inhibited by anisomycin, cycloheximide, chloramphenicol, tetracycline, streptomycin, erythromycin, and puromycin. |
| Localization | Found in the nucleus of eukaryotes and in the cytoplasm of prokaryotes. | Found in the cytoplasm of prokaryotes and on ribosomes on the endoplasmic reticulum in eukaryotes. |
FAQ
What is the main difference between transcription and translation?
The main difference between transcription and translation is their function in gene expression. Transcription is the process of synthesizing an RNA molecule from a DNA template, while translation is the process of decoding the mRNA sequence and synthesizing a protein.
Where does transcription occur, and where does translation occur?
Transcription occurs in the nucleus of eukaryotic cells, where DNA is transcribed into RNA. Translation, on the other hand, takes place in the cytoplasm, where mRNA is translated into a protein.
What is the purpose of transcription in gene expression?
The purpose of transcription is to convert the information encoded in DNA into an RNA molecule, specifically mRNA. This mRNA serves as a template for protein synthesis during translation.
How is RNA polymerase involved in transcription?
RNA polymerase is an enzyme that plays a crucial role in transcription. It binds to the DNA template strand and catalyzes the synthesis of a complementary RNA molecule by adding nucleotides according to the DNA sequence.
What is the role of ribosomes in translation?
Ribosomes are the cellular structures responsible for protein synthesis during translation. They consist of two subunits, large and small, which come together to form a functional ribosome. Ribosomes read the mRNA sequence and link amino acids together to form a polypeptide chain.
What are the starting molecules in transcription and translation?
In transcription, the starting molecule is the DNA template strand, which serves as a template for RNA synthesis. In translation, the starting molecule is the mRNA molecule, which carries the genetic code from DNA to the ribosome.
What is the end product of transcription, and what is the end product of translation?
The end product of transcription is an RNA molecule, which can be different types such as mRNA, tRNA, rRNA, or non-coding RNA. The end product of translation is a polypeptide chain, which folds into a functional protein.
How does the genetic code in DNA relate to transcription and translation?
The genetic code in DNA is transcribed into RNA during transcription. The sequence of nucleotides in DNA is translated into a complementary sequence of RNA bases (A, U, G, and C). This RNA sequence is then translated during protein synthesis to determine the sequence of amino acids in the protein.
What are the major steps involved in transcription?
The major steps involved in transcription include: pre-initiation, initiation, elongation, and termination. During pre-initiation, the RNA polymerase binds to the promoter region on the DNA. In initiation, the RNA polymerase begins synthesizing an RNA molecule. Elongation involves the continued synthesis of RNA along the DNA template. Termination occurs when the RNA polymerase reaches a termination sequence and releases the RNA molecule.
What are the major steps involved in translation?
The major steps involved in translation are: initiation, elongation, and termination. During initiation, the small ribosomal subunit binds to the mRNA, and the initiator tRNA binds to the start codon. Elongation involves the addition of amino acids to the growing polypeptide chain as the ribosome moves along the mRNA. Termination occurs when a stop codon is reached, signaling the release of the completed polypeptide chain and the disassembly of the ribosome.