When it comes to DNA replication and amplification, primers play a crucial role. Primers are short nucleotide sequences that provide a starting point for DNA synthesis. However, not all primers are created equal. There are two main types: RNA primers and DNA primers. Understanding the difference between RNA primers and DNA primers is essential for various molecular biology applications.
RNA primers are short strands of RNA, while DNA primers are short strands of DNA. This fundamental distinction sets the stage for their divergent characteristics and functions. RNA primers, typically composed of ribonucleotides, are used in DNA replication in living organisms. They are synthesized by a specialized enzyme called primase and provide a free 3′-OH group for DNA polymerase to extend from, initiating DNA replication. However, RNA primers are relatively unstable at higher temperatures due to the presence of hydroxyl groups on the ribose sugar.
On the other hand, DNA primers are synthetic oligonucleotides made up of deoxyribonucleotides. They are longer than RNA primers, ranging from 12 to 22 oligos, and are primarily used in DNA amplification techniques like Polymerase Chain Reaction (PCR). Unlike RNA primers, DNA primers remain a part of the newly synthesized DNA strand after replication or amplification. They are designed to target specific DNA sequences and are essential for the success of PCR experiments.
Furthermore, the synthesis of RNA primers relies on the action of primase enzyme, a specialized RNA polymerase, whereas DNA primers can be synthesized artificially using various methods like chemical synthesis or enzymatic amplification. RNA primers are typically single-stranded RNA molecules, while DNA primers consist of two single-stranded DNA molecules, namely forward and reverse primers, used for targeting different single-stranded DNA targets.
In summary, the difference between RNA primers and DNA primers lies in their composition, length, stability, function, and production methods. RNA primers are involved in DNA replication and have a shorter length, while DNA primers are utilized in DNA amplification techniques like PCR and are longer in length. Understanding these distinctions is crucial for selecting the appropriate primer type for specific molecular biology applications.
In the next section, we will explore the characteristics and specific roles of RNA primers and DNA primers in more detail, providing a comprehensive understanding of their respective functions and applications. Stay tuned!
What is RNA Primer?
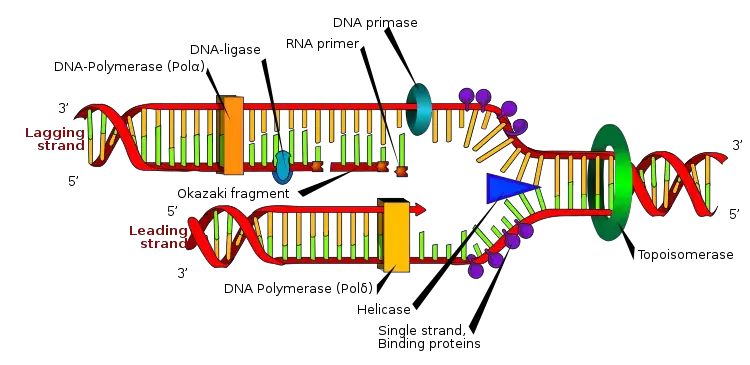
- An RNA primer is a short piece of RNA that serves as a starting point for DNA synthesis during DNA replication. It is essential because DNA polymerases, the enzymes responsible for copying DNA, require a pre-existing chain to add new nucleotides onto. Since DNA polymerases cannot initiate DNA synthesis on their own, RNA primers are synthesized by another enzyme called primase.
- During DNA replication, primase synthesizes a short RNA primer that is complementary to the DNA template strand. The RNA primer provides the necessary 3′-OH (hydroxyl) group for DNA polymerase to add nucleotides and extend the chain. DNA polymerase then extends the RNA primer by adding DNA nucleotides in a complementary manner to the template strand, resulting in the synthesis of a new DNA strand.
- After the DNA polymerase has completed the synthesis of the new DNA strand, the RNA primer is removed by another enzyme called DNA polymerase I or DNA polymerase δ. This process is known as primer removal or primer excision. The resulting gap is then filled with DNA nucleotides by DNA polymerase and sealed by another enzyme called DNA ligase, completing the replication process.
- Overall, RNA primers are crucial in DNA replication as they provide the initial point for DNA synthesis and allow the DNA polymerase to start building a new DNA strand.
What is DNA Primer?
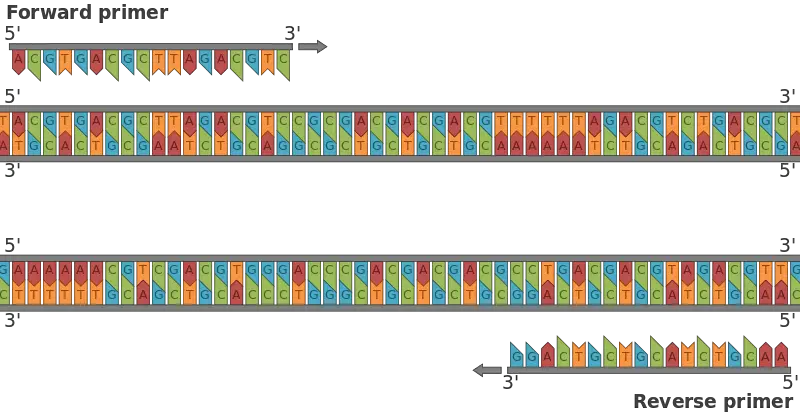
- A DNA primer is a short, single-stranded DNA fragment that plays a crucial role in laboratory techniques like polymerase chain reactions (PCR). PCR involves the hybridization of a pair of primers with a sample DNA, defining the specific region that will be amplified. This process generates millions of copies of the target DNA sequence in a very short period of time. DNA primers are also utilized in other experimental processes, such as DNA sequencing.
- Typically, DNA primers are around 18 to 25 nucleotides in length and can be synthesized in laboratories. The 3′ end of the primer serves as the starting point for DNA synthesis, while the 5′ end is often equipped with a fluorescent or radioactive tag. This tag facilitates the detection and analysis of the PCR product. The specificity of a primer is determined by its nucleotide sequence, which should be unique to the target DNA region being amplified.
- The efficiency of PCR amplification can be influenced by the GC content of the primer sequence. Primers with a high GC content tend to form stable duplexes with the template DNA, promoting efficient amplification. However, high GC content can also lead to the formation of secondary structures, such as hairpins, which can interfere with the amplification process.
- Additionally, modifications can be applied to DNA primers to enhance their performance or facilitate downstream analysis of the PCR product. For example, primers can be labeled with a fluorescent tag, enabling easier detection and purification of the amplified DNA fragments.
- In summary, DNA primers are short, single-stranded DNA fragments utilized in various laboratory techniques, especially PCR. They define the target region to be amplified, and their specificity and GC content influence the efficiency of amplification. Modifications and labeling can be employed to improve primer performance and aid in subsequent analysis of the PCR product.
Criteria to select the DNA primer
When selecting DNA primers for PCR amplification, several criteria should be considered:
- Length: The primer should typically be 12 to 20 nucleotides long for normal PCR amplification. This length is sufficient to provide specificity in binding to the target DNA sequence.
- GC Content: The GC content of the primer is an important factor. It is generally recommended to have a GC content of less than or between 45-50%. This range helps ensure optimal binding specificity and stability of the primer-template duplex.
- Hairpin Formation: Primers should have a low tendency to form hairpin structures, which are self-complementary sequences that fold back on themselves. High GC content can contribute to hairpin formation. Minimizing hairpin formation is crucial as it can interfere with primer binding and subsequent amplification.
- Annealing Temperature: The annealing temperature of the primer should be within a suitable range for PCR amplification. Typically, the annealing temperature is set between 55 to 65°C, depending on the specific primers and the target DNA sequence.
It is worth noting that when primers are used solely for amplification purposes, there is no need for proofreading or exonuclease activity. The DNA primers are designed to provide a substrate for DNA polymerase to initiate DNA synthesis during PCR.
Different polymerases are used for replication and amplification. The polymerase used in PCR amplification is stable at higher temperatures and lacks exonuclease activity. This allows it to withstand the denaturation step of PCR and continue DNA synthesis at elevated temperatures without degrading.
In contrast, the polymerase used in DNA replication works at lower temperatures and possesses exonuclease activity. This activity enables proofreading and removal of mismatched nucleotides during replication to ensure accurate DNA synthesis.
To summarize, the selection of DNA primers for PCR amplification involves considering their length, GC content, hairpin formation, and compatibility with the polymerase used in the amplification process.
What is the difference between an RNA primer and a DNA primer?
| Characteristics | RNA Primers | DNA Primers |
|---|---|---|
| Usage | DNA replication (in vivo) | DNA amplification during PCR (in vitro) |
| Length | 10 to 12 oligos | 12 to 22 oligos |
| Stability | Unstable at higher temperature | Stable at higher temperature |
| Exonuclease Activity | Removed after completion of replication | Remains a part of the newly synthesized DNA strand |
| Synthesis | Synthesized by DNA primase | Synthesized artificially |
| Molecule | Single-stranded RNA | Pair of single-stranded DNA (forward and reverse primers) |
| Nucleotide Composition | Adenine, guanine, cytosine, uracil | Adenine, guanine, cytosine, thymine |
| Composition | Consists of ribonucleotides | Consists of deoxyribonucleotides |
| Length | Typically 8-12 nucleotides | Typically 18-24 nucleotides |
| Function | Initiates DNA replication | Targets and amplifies specific DNA sequences |
| Stability | More prone to degradation due to hydroxyl groups | More stable due to absence of hydroxyl groups |
| Production | Synthesized by primase enzyme | Synthesized through chemical or enzymatic methods |
FAQ
What is the primary difference between an RNA primer and a DNA primer?
The main difference lies in their composition, where RNA primers are made of ribonucleotides, and DNA primers are made of deoxyribonucleotides.
How are RNA primers synthesized?
RNA primers are synthesized by a specialized enzyme called primase. Primase synthesizes short RNA sequences complementary to the DNA template.
What happens to RNA primers after DNA replication?
After DNA replication, RNA primers are removed by DNA polymerase and replaced with DNA nucleotides during a process called primer removal.
What is the role of an RNA primer?
RNA primers are crucial in DNA replication as they provide a starting point for DNA synthesis by DNA polymerase. They are later replaced with DNA nucleotides.
Can RNA primers be used in PCR?
No, RNA primers are not typically used in PCR. PCR primarily utilizes DNA primers due to their stability at higher temperatures required for amplification.
How do DNA primers differ from RNA primers in terms of stability?
DNA primers are more stable than RNA primers, especially at higher temperatures. DNA primers can withstand the denaturation step in PCR without degradation.
Can DNA primers be used for RNA amplification?
Yes, DNA primers can be used for amplifying RNA through reverse transcription PCR (RT-PCR) by first converting RNA to complementary DNA (cDNA).
Are there any differences in the synthesis process between DNA and RNA primers?
Yes, DNA primers are typically synthesized using automated DNA synthesis methods, whereas RNA primers are synthesized by enzymatic processes.
Do RNA primers have exonuclease activity?
No, RNA primers do not possess exonuclease activity. Exonuclease activity is typically associated with DNA polymerases used in DNA replication for proofreading.
Are there any differences in the annealing temperature between RNA and DNA primers?
Generally, RNA primers have lower annealing temperatures compared to DNA primers due to the higher stability of DNA-DNA duplexes compared to DNA-RNA duplexes.