What is cDNA?
- A cDNA is a functional part of DNA made from an RNA molecule. It is created using a special enzyme called reverse transcriptase.
- cDNA sequences as complementary DNA is different from the genomic DNA sequence, which only contain coding DNA sequences.
- cDNAsynthesis is an important first step for many molecular applications.
- Next generation sequencing (NGS), gene expression analysis, pathogen identification, and genetic testing by qPCR or NGS are just some examples of applications that require the conversion of RNA into cDNA as an early step.
- To get accurate and precise results from these applications requires high fidelity in cDNA library construction so that the cDNA libraries accurately represent the original RNA transcripts.
- When selecting a cDNA synthesis kit, two important considerations are the reverse transcriptase (RT) enzyme and the primer design strategy.
- Enzyme selection influences the transcription rate and reaction fidelity. If priming strategies are not optimized, they may result in a biased cDNA library that does not accurately reflect the entire transcriptome.
cDNA synthesis principle
- cDNA synthesis is an enzymatic process that involves the use of reverse transcriptase (RT) and forms complementary DNA (cDNA) from messenger RNA (mRNA). The whole process is technically known as reverse genetics.
- Naturally, it has existed in viruses like lentiviruses, human immunodeficiency virus 1 (HIV-1), murine leukemia virus, feline immunodeficiency viruses, etc., broadly classified as retroviruses.
- The polymerization property allows the reverse transcriptase enzyme to identify each nucleotide from the mRNA and form the complementary cDNA (cDNA) at the same time, by using the primer and incoming nucleotides.
- However, the in vivo process of cDNA synthesis is somewhat similar to the natural one, but differs in some ways. It is divided into two parts: First strand synthesis and second strand synthesis.
- We will learn about the in vitro process of complementing DNA synthesis in the next section.
cDNA synthesis steps
There are different steps for synthesis of cdna such as;
- Prepare sample
- Remove genomic DNA
- Select reverse transcriptase
- Prepare reaction mix
- Perform cDNA synthesis
cDNA synthesis protocol
1. Prepare sample
- The RNA template is used for cDNA production. The use of total RNA is common in cDNA production for downstream applications like RT-(q)PCR as well as specific kinds of RNAs (e.g. messenger RNA (mRNA) and smaller RNAs like miRNA) can be used in specific purposes like cDNA library building as well as miRNA profile profiling.
- In order to ensure the integrity of RNA, it is essential and requires special care for processing, extraction, storage, as well as experimental use.
- Most effective ways to avoid destruction of RNA are using gloves, pipetting with aerosol-barrier tips, using non-nuclease equipment and reagents, as well as disinfecting work areas.
- To purify and isolate RNA, various methods are available based on the sources (e.g. tissues, blood, cells and plants, etc.)) and the goals of the research.
- The main objectives of isolation workflows is to stabilize RNA molecules to reduce RNases and to increase yields by using proper storage and extraction techniques.
- Purification techniques that are optimally designed to eliminate endogenous substances, such as complex polysaccharides as well as humic acid from plant tissues, which inhibit enzyme activity and the most common inhibitors of reverse transcriptases like metal ions, salts the phenol and ethanol. After purification, RNA must be stored at -80°C with only a few freeze-thaw cycles.
2. Remove genomic DNA
- A small amount of genomic DNA (gDNA) can be co-purified with RNA. gDNA contamination can affect reverse transcription, and could result in false positives, a higher background or less detection for sensitive applications like the RT-qPCR.
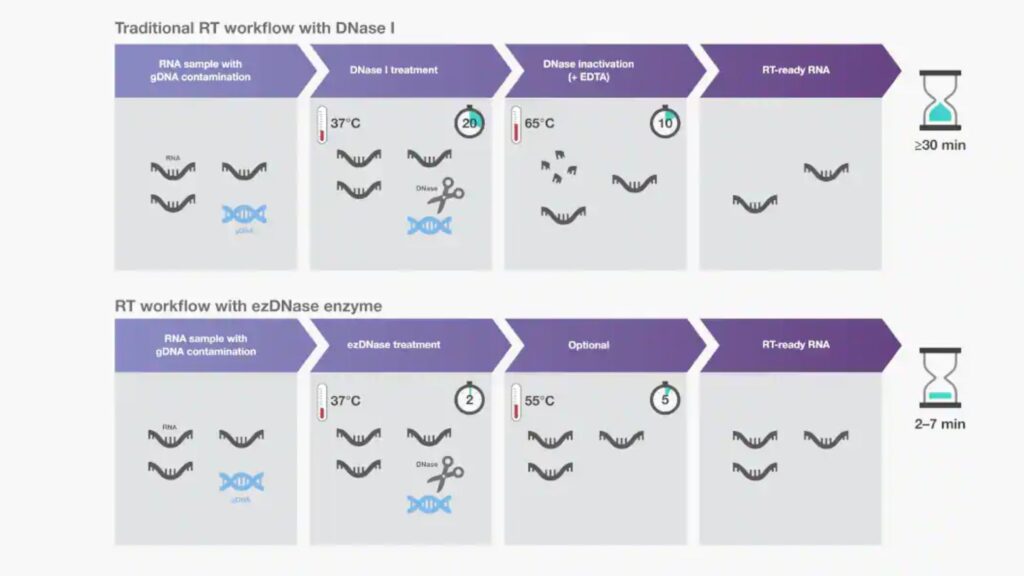
- The most common method for gDNA removal is to add DNase I to preparations of isolated DNA.
- DNase I needs to be eliminated prior to cDNA synthesizing because any remaining enzyme could degrade single-stranded DNA. Unfortunately, loss of RNA or damage could happen in DNase I inactivation treatment.
- As an alternative to DNase I, double-strand-specific DNases are available to eliminate contaminating gDNA without affecting RNA or single-stranded DNAs.
- Their thermolabile nature permits easy activation at a low temperature (e.g. 55 degrees Celsius) without negative impact.
- Such double-strand-specific, thermolabile DNases can be incubated with RNA for 2 min at 37degC prior to reverse transcription reactions to streamline the workflow
3. Select reverse transcriptase
- The majority of reverse transcriptases utilized in molecular biology originate from the gene Pol from the avian myeloblastosis virus (AMV) as well as Moloney Murine Leukemia Virus (MMLV).
- AMV reverse transcriptase enzyme was among the first enzymes discovered to facilitate cDNA synthesizing in the lab. It has a powerful RNase H activity that degrades the RNA of hybrids involving RNA and DNA which results in smaller CDNA pieces (<5 km).
- The reverse transcriptase of MMLV was an extremely popular choice because of its monomeric form that allowed for easier modification and cloning of the enzyme.
- While MMLV is more unstable than AMV reverse transcriptase MMLV reverse transcriptase has the capability of producing longer cDNA (<7 kb) with greater efficiency because of it having less RNase H activity.
- To further enhance cDNA synthesizing, MMLV reverse transcriptase has been designed to have less RNase H activities (i.e. mutation of the RNase H domain also known as RNaseH-) with a more temperature stability (up up to 55degC) and improved processing efficiency (65 times more efficient).
- These characteristics result in an increase in cDNA size and production, greater sensitivity, better resistance to inhibitors and more rapid reaction time.
Common reverse transcriptases and their attributes.
| AMV reverse transcriptase | MMLV reverse transcriptase | Engineered MMLV reverse transcriptase (e.g., Invitrogen SuperScript IV Reverse Transcriptase) | |
|---|---|---|---|
| RNase H activity | High | Medium | Low |
| Reaction temperature (highest recommended) | 42°C | 37°C | 55°C |
| Reaction time | 60 min | 60 min | 10 min |
| Target length | ≤5 kb | ≤7 kb | ≤12 kb |
| Relative yield (with challenging or suboptimal RNA) | Medium | Low | High |
4. Prepare reaction mix
- In addition to enzymes and primers, the primary reactions components used to reverse transcription are templates for RNA (pre-treated to eliminate genomic DNA) buffer, dNTPs, buffer, DTT, RNase inhibitor and RNase-free water.
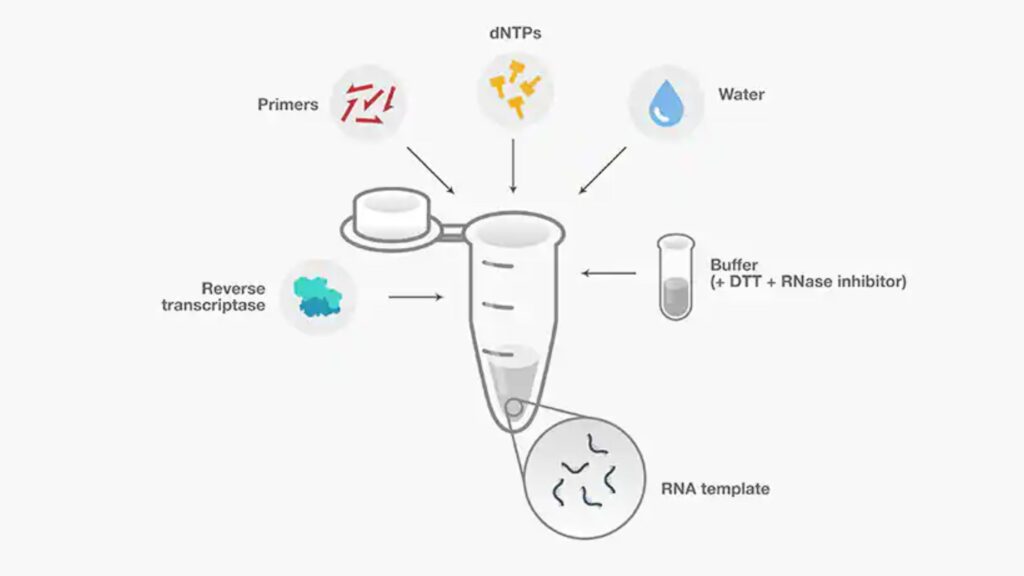
| Component | Key features |
|---|---|
| RNA template | It is crucial to ensure the integrity of RNA and requires extra security measures during extraction, processing storage, and research application (see step 1):Total the RNA that is routinely utilized in cDNA synthesizing for downstream applications like RT-(q)PCRMessenger (mRNA) (mRNA) and smaller RNAs like miRNA) can be used in specific applications, such as cDNA library building and miRNA profiling |
| Reaction buffer | Maintains a balanced pH and Ionic force for reaction.The buffer supplied could also contain additives that improve the effectiveness in reverse transcription. |
| dNTPs | Generally, should be at 0.5-1 mM each, preferably at equimolar concentrationsHigh-quality dNTPs, freshly diluted, are recommended to ensure proficient reverse transcription |
| DTT | Reducer agent, which is often used to maximize enzyme activity.Reaction effectiveness may be affected in the event that DTT and other ingredients disperse thus, reactions components must be dissolvable and thoroughly mixed. |
| RNase inhibitor | Most often, they are included in the buffer for reaction and added in the reverse transcription process to prevent degradation of RNA. They could be: Co-purified in isolationIntroduced in the process of setting up of well-known RNases are available, and suitable RNase inhibitors must be selected in accordance with their mechanism of action and the requirements for the reaction. |
| Water | Eliminate any RNases by using:Nuclease-free from a commercial sourceDEPC (diethylpyrocarbonate)-treated waterContaminating RNases cannot be removed by simple filtration, and autoclaved water is not adequate because RNases are heat stable. |
5. Perform cDNA synthesis
- Reverse transcription reactions require three major processes: primer annealing DNA polymerization as well as enzyme activation.
- The time and temperature of these steps varies based on the primer used, the target reverse transcriptase and target employed.
- The crucial phase is DNA polymerization. In this process, the duration and temperature of reaction may differ based on the primer selection and reverse transcriptase that is used.
- If you’re using random hexamers we recommend that you incubate this reverse transcription process at room temperature (~25 degree Celsius) for 10 minutes after adding the enzyme to increase the primers.
- There are different thermal stability levels which determines the most optimal temperature for polymerization for each.
- Utilizing a thermostable reverse transcription use allows for a higher temperature of reaction (e.g. 50degC) which helps remove RNA that has a high GC or secondary structures, but without affecting the enzyme’s activity.
- In the presence of such enzymes the high temperature of incubation could cause an improvement in cDNA production, size and also in the representation.
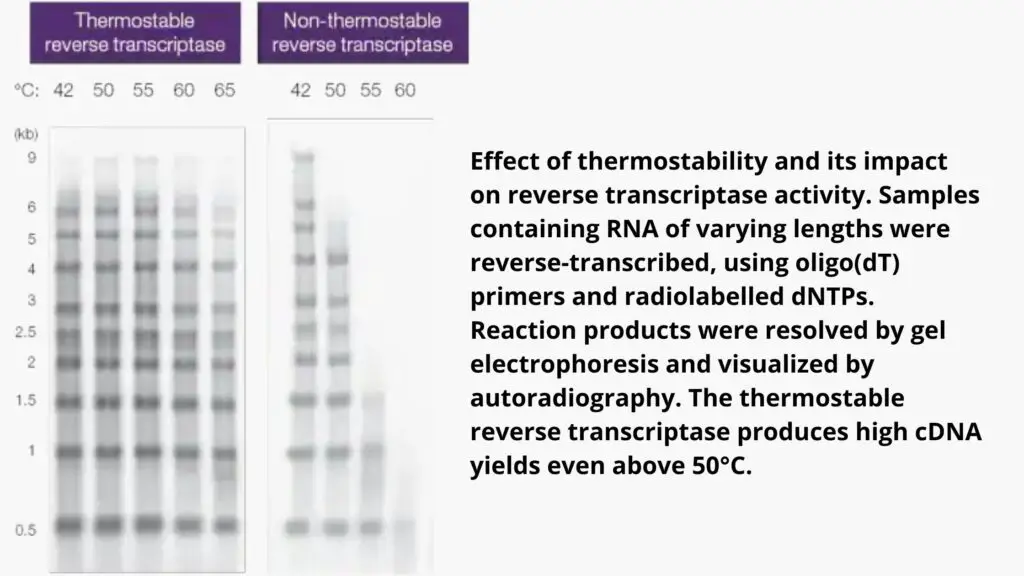
The duration of polymerization depends on the processivity of a reverse transcriptase which is the quantity of nucleotides included in a single binding. For instance, a wild-type reverse transcriptase that has low processivity typically requires more than 60 minutes to synthesize the cDNA. However reverse transcriptase engineered with high processivity could require less than 10 minutes to produce a 9kb DNA.
Troubleshooting tips
- Reduce the number of freeze-thaw cycles on the RNA samples in order to avoid degradation.
- The RNA is stored in an EDTA-buffered solution to reduce the nonspecific cleavage that occurs by nucleases which are cofactors of metal ions.
- Use water that is certified nuclease-free or treated with DEPC (diethylpyrocarbonate) to ensure the absence of RNase.
- Examine the quality of RNA using microfluidics or gel electrophoresis.
- The amount of contaminants that are removed in the process of purifying RNA (e.g., SDS, EDTA) could hinder DNase functions, so make a second attempt to extract the RNA using alcohol, clean the pellet with 75% ethanol and then rinse it with nuclease-free water.
- Choose the gDNA removal method that is not detrimental to the integrity of RNA. If you are using ezDNase, extend the treatment to 5 min of incubation at 37oC.
- DNase is a very sensitive enzyme. Therefore when you’re applying DNase treatments, our best way to go is to mix it gently pipetting upwards and downwards vs. vortexing.
- The most efficient method for removing DNase is to do the phenol/chloroform extraction, or employ spin columns.
- If you begin with a small amount of RNA, be sure you select an reagent for reverse transcription that has high linearity over the entire range of RNA inputs, to ensure that RNA with low abundance can be accurately quantified.
- When working with RNA which has been degraded or with residual salts and inhibitors, think about a reverse transcriptase enzyme that is able to effectively work with RNA that has been degraded or is intolerant to salts, as well as carryover biological inhibitors as well as extraction agents.
- The qRT-PCR is an extremely sensitive instrument for analyzing RNA. Since the PCR amplifies the target, any errors are amplified. Thus, the degree of variability should be minimized when it is possible. Thinking about the possibility of a “master mix” or mix of reaction reagents is recommended for making multiple reactions to reduce sample-to-sample as well as well-to-well variability and increase reproducibility.
- Mix the reagents thoroughly to dissolve DTT and salts that could have precipitated.
- If your RNA sample has a significant GC quantity or another secondary structural component, reduce the growth of hairpin sequences through reverse transcription at the higher temperature (e.g. 50oC). Think about using a thermostable reverse transcriptase, which can withstand higher temperatures for reaction.
- A problem with the design of primers can be prevented by using reverse transcription at a high temperature, which can enhance the specificity of the primer binding. You can also use a thermostable reverse transcriptase.
cDNA synthesis ppt
References
- https://international.neb.com/applications/dna-amplification-pcr-and-qpcr/rt-pcr-and-cdna-synthesis/cdna-synthesis
- https://www.bio-rad.com/webroot/web/pdf/lsr/literature/4106228.pdf
- https://geneticeducation.co.in/the-process-of-cdna-synthesis-and-cdna-library-preparation/
- https://www.takarabio.com/products/mrna-and-cdna-synthesis
- https://www.bio-rad.com/featured/en/cdna-synthesis.html
- https://pcrbio.com/row/applications/cdna-synthesis/
- https://international.neb.com/protocols/0001/01/01/first-strand-cdna-synthesis-e6300