The virus uses the host’s complete system for protein translation because they lack translational machinery. Hence, the Baltimore classification system is based on this machinery how viruses use the host mechanism. Messenger RNA (mRNA) is the main focus of this classification system.
In the beginning, there were six classes of viruses in Baltimore classification system but later a new class was added to accommodate the gapped DNA genome of Hepadnaviridae (hepatitis B virus).
Baltimore classification System
- In 1971, a Nobel Prize-winning virologist David Baltimore first introduced the Baltimore Classification of the virus. The Baltimore classification system is considered as the most commonly used system of virus classification.
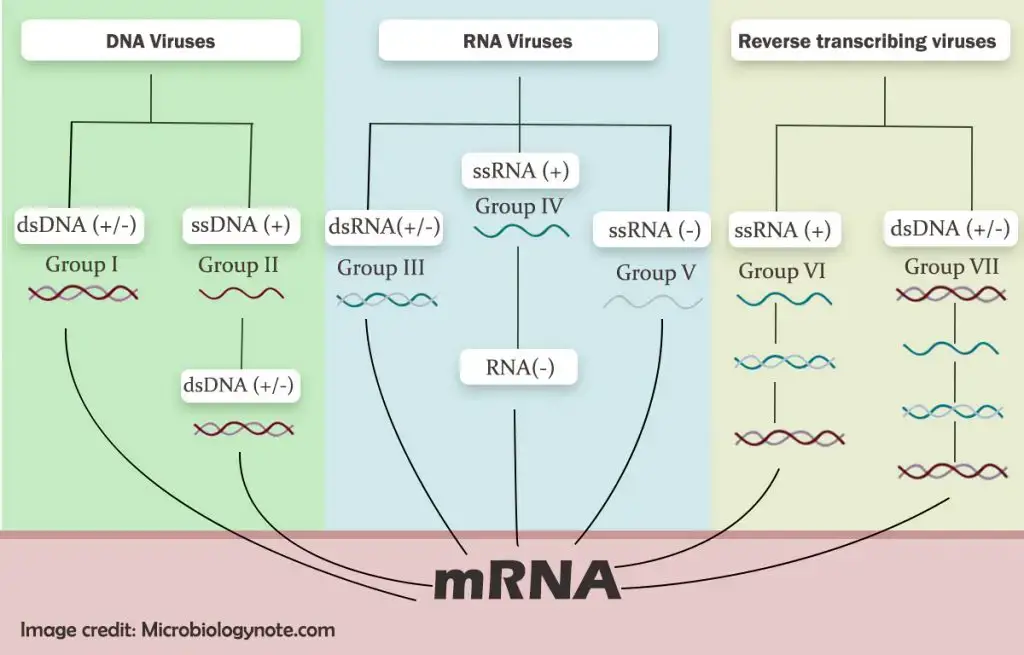
- This classification system divides the viruses into seven groups based on their manner of messenger RNA (mRNA) synthesis. In this classification, the viruses were organized based on their manner of mRNA synthesis during the replication cycle of virus.
- The Baltimore classification system focuses on a few important characteristics of nucleic acid such as either it made up of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA); genome strandedness such as either single- or double-stranded; sense of the genome such as either positive or negative sense.
Baltimore classification with Chart
There are seven classes of virus in Baltimore classification such as;
DNA virus
The DNA viruses are divided into two groups such as double-stranded DNA (dsDNA) viruses, and single-stranded DNA (ssDNA) viruses.
1. Group I: double-stranded DNA viruses
- All the viruses of group I contain double-stranded DNA (dsDNA).
- They synthesized their mRNA in a three-step process.
- In the 1st step of mRNA synthesis, a transcription preinitiation complex binds at the transcription site on DNA, and enable the requirement of host RNA polymerase.
- In the 2nd step, when the RNA polymerase is supplied it synthesis mRNA strands from the negative strand by using it as a template.
- In the last step of mRNA synthesis, the RNA polymerase reaches a specific signal, known as the polyadenylation site, and terminates the transcription processes.
- They use different methods for their genomic replication such as bidirectional replication, rolling circle mechanism, etc.
- There is a subdivision of dsDNA viruses between those replicate in the nucleus and uses the host’s cell machinery for transcription and replication, and those are replicating in the cytoplasm by acquiring their own means of executing transcription and replication.
The dsDNA is divided into three realms such as;
- Duplodnaviria:
- These are dsDNA viruses.
- They belong to two groups such as tailed bacteriophages in Caudovirales and herpesviruses in Herpesvirales.
- Monodnaviria:
- These are the members of Papovaviricetes and all of them are dsDNA viruses.
- They consist of two groups such as papillomaviruses and polyomaviruses.
- Varidnaviria:
- Adenoviruses, giant viruses, and poxviruses are included within this realm.
Example of Group I Viruses: Herpesviridae, Adenoviridae, and Papovaviridae.
2. Group II: single-stranded DNA viruses
- This group of viruses contains a single-stranded DNA (ssDNA) genome. They follow the same transcription mechanism as the dsDNA viruses do.
- Most of them contain circular genomes, which are replicated by rolling circle replication (RCR). The RCR is initiated when the positive strand is cleaved by an endonuclease enzyme and as a result, the DNA polymerase starts to use the negative strand as a template for replication.
- Parvoviruses have linear ssDNA genomes, they use rolling hairpin replication (RHR) for the replication of their genome.
Example of Group II Viruses: Anelloviridae, Circoviridae, and Parvoviridae.
RNA Virus
The RNA viruses have three groups such as double-stranded RNA (dsRNA) viruses, positive-sense single-stranded RNA (+ssRNA) viruses, and negative-sense single-stranded RNA (-ssRNA) viruses. They are divided into the kingdom Orthornavirae in the realm of Ribavirin.
3. Group III: double-stranded RNA viruses
- Viruses of this group contain double-stranded RNA (dsRNA).
- After entering the host body the RNA-dependent RNA polymerase (RdRp) transcribed the dsRNA genome into mRNA, later this transcribed mRNA is used for the translation or replication.
Example of Group III Viruses: Reoviridae and Birnaviridae.
4. Group IV: positive-sense single-stranded RNA viruses
- This group contains only those viruses that have a positive-sense single-stranded RNA (+ssRNA) genome.
- They don’t require transcription for translation because the genome functions as mRNA.
- Intermediates of dsRNA, referred to as replicative intermediates, are made within the strategy of copying the genomic RNA.
- A number of, full-length RNA strands of destructive polarity (complimentary to the positive-stranded genomic RNA) are fashioned from these intermediates, which can then function templates for the manufacturing of RNA with constructive polarity, together with each full-length genomic RNA and shorter viral mRNAs.
Example of Group IV Viruses: Coronaviridae, Flaviviridae, Astroviridae, and Picornaviridae.
5. Group V: negative-sense single-stranded RNA viruses
- Viruses of this group contain negative sense, single-stranded RNA (-ssRNA) genome.
- Their sequence is complementary to the mRNA.
- The positive sense mRNA is directly transcribed from the negative sense genome.
Example of Group V Viruses: Orthomyxoviridae, Paramyxoviridae, and Rhabodviridae.
Reverse transcribing viruses
Their genome is made of either DNA or RNA and replicates through the reverse transcription. There are two groups of reverse transcribing viruses such as single-stranded RNA-RT (ssRNA-RT) viruses, and double-stranded DNA-RT (dsDNA-RT) viruses. They are divided into kingdom Pararnavirae in the realm Riboviria.
6. Group VI: single-stranded RNA viruses with a DNA intermediate in their life cycle
- Viruses of this group contain a (positive-sense) single-stranded RNA genome that has a DNA intermediate ((+)ssRNA-RT) in its replication cycle.
- They convert their linear genome into dsDNA by using reverse transcription. After that the dsDNA is transferred into the host nucleus and then inserted into the host genome. This genome is used for the synthesis of mRNA.
Example of Group VI Viruses: retroviruses such as HIV, as well as Metaviridae and Pseudoviridae.
7. Group VII: double-stranded DNA viruses with an RNA intermediate in their life cycle
- Viruses of this group contains a double-stranded DNA genome that has an RNA intermediate (dsDNA-RT) in its replication cycle.
- They contain partial dsDNA genomes and produce ssRNA intermediates which act as mRNA, but also the reverse transcriptase enzyme again converts this ssRNA into dsDNA which is necessary for genome replication.
Example of Group VII Viruses: Hepatitis B virus
Table of Baltimore Classification System
| Name of Groups | Nucleic acid types | Manners of mRNA Production | Example |
| Group I | Double-stranded DNA | mRNA is transcribed directly from the DNA template | Herpes simplex (herpesvirus) |
| Group II | Single-stranded DNA | DNA is converted to double-stranded form before RNA is transcribed | Herpes simplex (herpesvirus) |
| Group III | Double-stranded RNA | mRNA is transcribed from the RNA genome | Childhood gastroenteritis (rotavirus) |
| Group IV | Single stranded RNA (+) | Genome functions as mRNA | Common cold (pircornavirus) |
| Group V | Single stranded RNA (-) | mRNA is transcribed from the RNA genome | Rabies (rhabdovirus) |
| Group VI | Single stranded RNA viruses with reverse transcriptase | Reverse transcriptase makes DNA from the RNA genome; DNA is then incorporated in the host genome; mRNA is transcribed from the incorporated DNA | Human immunodeficiency virus (HIV) |
| Group VII | Double stranded DNA viruses with reverse transcriptase | The viral genome is double-stranded DNA, but viral DNA is replicated through an RNA intermediate; the RNA may serve directly as mRNA or as a template to make mRNA | Hepatitis B virus (hepadnavirus) |
Why Baltimore Classification System is based on mRNA?
Baltimore classification systems are based upon the fundamental role of the translational apparatus and put mRNA at the heart and described the steps to create mRNA using DNA or the RNA genomes. Viral infections can replicate DNA or DNA, produce DNA from DNA, or in reverse, but do not have the complete mechanism to create proteins, and for this they depend on ribosomes of host cells. Host cells, on the other however, can produce proteins through +mRNA strands. No matter the genome-specific nature of viruses, all viruses have to synthesize viral + mRNAs in order to generate viral proteins”no exceptions to date”.
How do you identify positively (+) or Negative (-) strand?
The mRNA which can serve as the template for protein synthesis is defined as a positive (+) strand . A DNA strand that has the same polarity is known as “the (+) strand. DNA and RNA strands that are complementary with those of the (+) strand are known as negative (-) Strands.
References
- https://en.wikipedia.org/wiki/Baltimore_classification
- https://www.news-medical.net/life-sciences/The-Baltimore-Classification-System.aspx
- https://viralzone.expasy.org/254
- https://www.slideshare.net/theophilus74/baltimore-classification-of-viruses-presentation
- https://byjus.com/neet-questions/what-is-the-baltimore-classification/
- https://www.bionity.com/en/encyclopedia/Virus_classification.html
- https://microbeonline.com/baltimore-system-classifications-viruses/
The information is fully provided. it has helped me in getting a wider scope on Baltimore classification
nice notes for pharmacy and puc i recomended for this