What Is Bacterial Transformation?
- Bacterial transformation is a natural process where free DNA from a donor bacterium is absorbed by a recipient bacterium from the surrounding environment. This uptake of DNA often leads to the expression of new traits in the recipient, marking it as a transformant. This process is distinct in that it does not require the donor cell to be alive, only the presence of free DNA in the extracellular space.
- This phenomenon occurs when certain bacterial species release their DNA into the environment, often as a response to environmental stress. These released DNA fragments can then be taken up by competent cells—bacteria with the ability to incorporate external genetic material. These cells regulate gene uptake as part of the natural transformation mechanism, responding dynamically to environmental cues.
- Transformation plays a critical role in genetic exchange and adaptation, as it allows for the horizontal transfer of genetic material. It is particularly valuable in laboratory settings, where it is widely used to introduce modified or synthetic DNA into bacterial cells for research and biotechnological applications. Through this method, DNA segments ranging from one to tens of kilobases can be successfully transferred and expressed in recipient cells.
Bacterial Transformation Definition
Bacterial transformation is the process by which a bacterium takes up free DNA from its environment and incorporates it into its genome, potentially expressing new traits.
Principle of Bacterial Transformation
- The principle of bacterial transformation relies on the ability of bacteria to take up free DNA from their environment and incorporate it into their own genome or plasmids. This process is naturally utilized by certain bacteria and can also be induced in laboratory settings for research and genetic engineering purposes.
- The key factor for successful transformation is the competence of the recipient cell, which refers to its ability to take up and integrate naked DNA. Naturally competent bacteria often release DNA during the late stationary phase through cell autolysis, making it available for uptake by other cells. In a laboratory environment, bacteria such as Escherichia coli can be artificially made competent using chemical treatments like calcium chloride, or physical methods such as electroporation or heat shock. These methods enhance the permeability of the bacterial cell wall, enabling the entry of donor DNA.
- Once inside the cell, the donor DNA can integrate into the host genome through homologous recombination or remain as part of an autonomously replicating plasmid. If a plasmid is involved, it can carry specific genes, such as those encoding antibiotic resistance or enzymes for metabolic functions, which are later expressed in the transformant. This allows researchers to identify successful transformations by growing bacteria on media containing antibiotics or other selective agents.
- Transformation also enables the introduction of DNA containing selectable markers, ensuring that only cells that have successfully incorporated the new genetic material survive under selective conditions. This principle makes bacterial transformation a powerful tool for studying gene function and creating genetically modified organisms.
Types of Bacterial Transformation
Bacterial transformation can occur naturally or be induced artificially. Both forms allow bacteria to take up foreign DNA, but they differ in the way competence is achieved and in their underlying processes.
1. Natural Transformation
- Competence: Some bacteria naturally possess the ability to take up DNA from their surroundings without external intervention.
- Process: The DNA from the environment is absorbed directly into the bacterial cell, typically through specialized channels in the cell membrane.
- Species-Specific: Not all bacteria are naturally competent. This process is common in species like Streptococcus pneumoniae and Neisseria gonorrhoeae.
- Genetic Exchange: In natural transformation, genetic material is often incorporated into the bacterial genome, allowing for genetic diversity and adaptation.
2. Artificial Transformation
- Induced Competence: Unlike natural transformation, artificial transformation requires the bacterial cell’s competence to be induced through external techniques.
- Heat Shock Method: Cells mixed with DNA are exposed to a sudden increase in temperature (e.g., 42°C), which helps the cell take up the DNA.
- Electroporation: Cells are subjected to an electric field that temporarily makes the cell membrane permeable, allowing DNA to enter.
- Controlled Environment: Artificial transformation is commonly used in laboratories where specific genetic modifications are required.
- Applications: This form is widely employed in genetic engineering, where foreign DNA, such as plasmids, is introduced into bacterial cells to study gene function or produce recombinant proteins.
Steps of Bacterial Transformation
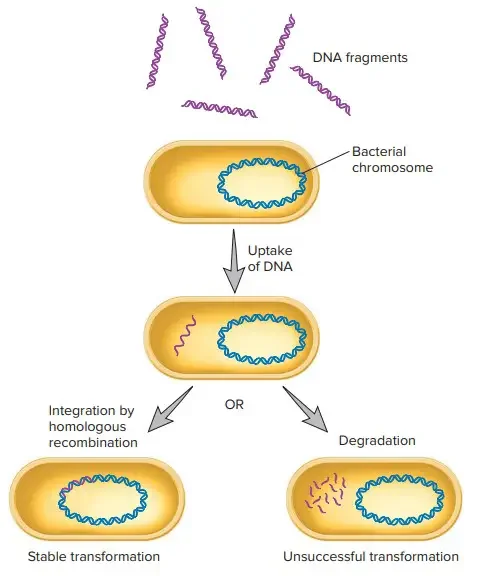
Bacterial transformation is a critical process used to introduce foreign DNA into bacterial cells, enabling genetic manipulation for research and industrial applications. The process involves four main stages: competence development, DNA binding, DNA uptake and processing, and integration into the host genome.
Key Steps of Bacterial Transformation
- Development of Competence
Bacteria must enter a competent state to take up DNA from their surroundings. Competence can occur naturally in some bacterial species or be artificially induced.- Heat Shock Method: Cells mixed with DNA are chilled at 0°C, then rapidly exposed to 42°C, facilitating DNA uptake.
- Electroporation: The cell-DNA mixture is subjected to a brief high-voltage electric pulse, temporarily creating pores in the membrane for DNA entry.
- DNA Binding to Cell Surface
Once competence is achieved, double-stranded DNA (dsDNA) from lysed cells binds to receptors on the bacterial surface. This interaction is non-specific, meaning DNA from various sources, even outside the species, can bind. - Processing and Uptake of DNA
Membrane-bound endonucleases cleave the bound dsDNA into smaller fragments. The resulting single-stranded DNA (ssDNA) is channeled into the bacterial cell through specialized translocation proteins embedded in the membrane. - Integration of DNA into the Host Chromosome
The single-stranded DNA that enters the cell undergoes homologous recombination, replacing a corresponding segment of chromosomal DNA. This process requires extensive sequence similarity between the incoming DNA and the host genome. - Transformation with Plasmids
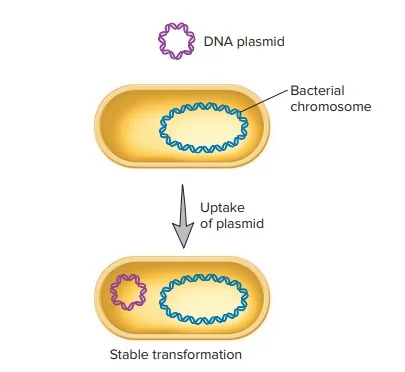
If the introduced DNA is in plasmid form, the plasmid does not require chromosomal integration. Instead, it remains extrachromosomal and replicates independently. Bacteria transformed with plasmids can be selected by growing them on media containing specific antibiotics, as plasmids often carry antibiotic resistance genes.
Bacterial Transformation Protocols
Bacterial transformation protocols are essential for introducing foreign DNA into bacterial cells. Two common methods used are the chemical transformation of competent cells and electroporation of electrocompetent cells. Each protocol involves specific steps and conditions to ensure successful DNA uptake and propagation in bacterial cultures.
Requirements Before Starting Transformation
Before initiating the bacterial transformation process, it’s essential to prepare various components to ensure that the procedure runs smoothly and effectively. Here’s what needs to be set up:
- Prepare LB Agar Plates:
- Prepare fresh LB agar plates or use pre-poured ones. If using pre-poured plates, ensure they are warmed to 37°C before use.
- Add Antibiotics:
- Incorporate the appropriate antibiotic based on the plasmid DNA used in the transformation. This ensures selective growth of transformed cells.
- Blue/White Screening Setup (Optional):
- If performing blue/white screening to identify recombinants, add the following to the LB agar:
- 1 mM IPTG (for inducing expression of lacZ gene)
- 300 µg/mL S-Gal or 40 µg/mL X-Gal (for colorimetric detection of recombinants)
- 500 µg/mL ferric ammonium citrate (to visualize the reaction).
- If performing blue/white screening to identify recombinants, add the following to the LB agar:
- Prepare Electroporation Chamber (If Using Electrocompetent Cells):
- Ensure the electroporation chamber is placed on ice to maintain the cells at low temperatures.
- Water Bath Setup:
- Heat the water bath to 37°C to prepare for the heat shock step in the transformation protocol.
- SOC Medium Preparation:
- Warm the sterile SOC medium to room temperature (or 20-25°C in a water bath). This medium will aid in cell recovery post-transformation.
Protocol for Transformation Using Chemically Competent Cells
- Prepare Cells:
- Retrieve the necessary number of tubes from the -70°C freezer and place them on wet ice.
- Allow cells to thaw for approximately 5 minutes, gently tapping the tubes to achieve a uniform suspension.
- Control DNA (Optional):
- For a control reaction, add 1 µL (10 ng) of pUC19 control DNA to one tube, mix by gentle tapping, and place the tube on ice.
- Add DNA to Cells:
- For transformation, add 1 ng to 50 ng of purified plasmid DNA directly to the competent cells in the remaining tubes. Gently mix and place the tubes on ice.
- Incubate on Ice:
- Incubate the cells on ice for 30 minutes to allow the DNA to interact with the cells.
- Heat Shock:
- Transfer the cells to a 37°C water bath for exactly 45 seconds to facilitate the uptake of DNA.
- Return to Ice:
- Immediately place the cells on ice for 2 minutes to stabilize the transformation.
- Recovery and Incubation:
- Add SOC medium to each tube to help the cells recover.
- Transfer the cells to sterile polypropylene tubes and loosen the caps to promote aeration.
- Incubate the cells in a shaker incubator at 37°C and 225-250 rpm for 1 hour.
- Plating:
- Pipette 10-100 µL of the transformed cell suspension onto LB agar plates containing the appropriate selection antibiotic.
- Spread evenly using a sterile spreader.
- Incubate the plates at 37°C overnight.
- Colony Selection:
- After overnight incubation, select colonies for further culture.
- Isolate plasmid DNA from each culture and digest using restriction enzymes for analysis via gel electrophoresis.
Protocol for Transformation Using Electrocompetent Cells
- Prepare Cells:
- Remove the required number of tubes from the -70°C freezer and place on wet ice.
- Thaw the cells for 5 minutes and tap the tubes to ensure a uniform suspension.
- SOC Medium:
- Add SOC medium to culture tubes, ensuring the final volume (cells and medium) is 1000 µL per transformation reaction.
- Allow the medium to reach room temperature.
- Prepare Cuvettes:
- Place sterile 1 mm cuvettes and microcentrifuge tubes on ice, one set per transformation.
- Transfer Competent Cells:
- Transfer competent cells into chilled microcentrifuge tubes. For each reaction, use 40 µL from an 80 µL package or 50 µL from a 100 µL package.
- Control DNA (Optional):
- Add 1 µL of 1-5 fold diluted pUC19 control DNA into one tube.
- DNA Dilution:
- Dilute the DNA or ligation mixture 1-5 fold in TE buffer. Add this mixture to the microcentrifuge tubes containing the competent cells.
- Electroporation:
- Pipette the DNA-cell mixture into a chilled 1 mm cuvette.
- Set the electroporator to a field strength of 25 kV/cm for 6 ms.
- Electroporate the cells and immediately transfer them into tubes containing SOC medium.
- Recovery and Incubation:
- Incubate the cells in a shaker incubator at 37°C and 225-250 rpm for 1 hour.
- Plating:
- Pipette 10-100 µL of the transformed cell suspension onto LB agar plates with the appropriate selection antibiotic.
- Spread evenly using a sterile spreader.
- Incubate the plates at 37°C overnight.
- Colony Selection:
- After incubation, select colonies for further culture.
- Isolate plasmid DNA from each culture and perform restriction enzyme digestion.
- Analyze the DNA using gel electrophoresis to verify successful transformation.
Applications of Bacterial Transformation
Bacterial transformation plays a critical role in various scientific and medical fields. It not only aids in understanding bacterial genetics but also has practical applications in biotechnology, medicine, and research.
- Gene Cloning:
- Bacteria can be transformed with recombinant plasmids, allowing them to replicate specific DNA sequences.
- This method is essential for producing large quantities of genes for further analysis or manipulation, making it a cornerstone of molecular cloning.
- Protein Production:
- Once transformed, bacteria can express proteins encoded by the introduced genes.
- This is crucial for the production of proteins such as enzymes, hormones, and biopharmaceuticals, with significant implications in medicine and industry.
- Acquisition of New Traits:
- Transformation allows bacteria to acquire new traits like antibiotic resistance or enhanced metabolic functions.
- These traits increase the bacteria’s ability to survive in changing environments, giving them a survival advantage.
- Genetic Mixing:
- Through transformation, bacteria can exchange genetic material, leading to genetic mixing among populations.
- This genetic exchange facilitates the emergence of new phenotypes and boosts the bacteria’s adaptability to environmental stressors.
- Genome Stability:
- Transformation can also serve a protective function, helping bacteria stabilize their genomes by removing harmful mobile genetic elements (MGEs).
- This process enhances bacterial resistance to parasitic infections and contributes to genome integrity.
- Genetic Engineering:
- Transformation is a key tool in genetic engineering, enabling the creation of genetically modified organisms (GMOs).
- These techniques are widely used in agriculture, where they help develop crops with desirable traits, and in research for studying gene functions.
- Synthetic Biology:
- Bacterial transformation also supports the field of synthetic biology, which involves designing and constructing new biological parts, devices, and systems.
- These engineered organisms can be applied in innovative medical and environmental applications, pushing the boundaries of biological research.
Examples of Bacterial Transformation
There are a few prominent examples of this process that have been studied extensively, each with distinct characteristics.
- Streptococcus pneumoniae:
- The first example of bacterial transformation was observed in Streptococcus pneumoniae.
- This species can transform from smooth (capsule-positive) colonies to rough (capsule-negative) colonies.
- The transformation occurs when the bacterial cells take up DNA from dead cells, leading to changes in their capsule production.
- This phenomenon was critical in understanding genetic exchange in bacteria, laying the groundwork for later studies on bacterial genetics.
- Neisseria species:
- Neisseria species, such as Neisseria gonorrhoeae and Neisseria meningitidis, exhibit a form of transformation where they specifically take up DNA from their own species.
- The process relies on species-specific recognition, ensuring that the DNA incorporated is compatible with the bacteria’s own genome.
- This mechanism helps maintain genetic integrity within the species, preventing the uptake of foreign DNA that could be harmful or incompatible.
- Haemophilus influenzae:
- Similar to Neisseria, Haemophilus influenzae also demonstrates species-specific DNA uptake.
- This transformation process is critical for the bacterium’s adaptability, enabling it to acquire genetic material that can enhance its survival, such as antibiotic resistance genes.
- Bacillus subtilis:
- Bacillus subtilis is another well-known example of a bacterium capable of natural transformation.
- This species can take up DNA from the environment, a feature that has been crucial in studies of bacterial adaptation and genetic diversity.
- Natural transformation in B. subtilis plays a role in the bacteria’s ability to evolve and adapt to changing environmental conditions.
Comparison of Transformation and Transfection
| Transformation | Transfection |
|---|---|
| Definition | Uptake and incorporation of foreign DNA into bacterial cells |
| Organisms | Primarily bacteria |
| Method of Introduction | Natural or artificial transformation methods (e.g., heat shock or electroporation) |
| Cellular Uptake | DNA is taken up by bacteria through a series of steps, including DNA binding, uptake, and integration into the genome |
| DNA Uptake Mechanism | Natural competence or artificial induction of competence |
| Genetic Modification | Incorporation of foreign DNA into the bacterial genome, leading to heritable changes |
| Applications | Genetic engineering, gene function studies, antibiotic resistance studies in bacteria |
| Host Range | Bacterial species with natural competence or those made artificially competent |
| Efficiency | Variable, depends on bacterial strain, competence conditions, and DNA characteristics |
| Control Over Genetic Material | May involve introducing specific DNA fragments or plasmids into bacteria |
| Challenges | Transformation efficiency, selection of transformants, and optimization of conditions |
FAQ
What is bacterial transformation?
Bacterial transformation is the process by which bacteria take up and incorporate foreign DNA into their genome, either naturally or through laboratory techniques.
How does natural transformation occur in bacteria?
Natural transformation occurs when competent bacteria uptake and integrate free DNA fragments from their environment through a series of molecular processes, including DNA binding, uptake, and integration by recombination.
What is the difference between natural and artificial transformation?
Natural transformation is a spontaneous process that occurs in certain bacterial species, while artificial transformation is a controlled laboratory technique that involves the intentional introduction of foreign DNA into bacteria.
What are the steps involved in bacterial transformation?
The steps of bacterial transformation include the development of competence, binding of DNA to the cell surface, processing and uptake of free DNA, and integration of the DNA into the chromosome through recombination.
How is competence developed in bacteria for artificial transformation?
Competence can be artificially induced in bacteria through methods like heat shock treatment or electroporation. Heat shock involves subjecting the cell-DNA mixture to a temperature shift from ice to 42°C, while electroporation uses a brief high-voltage electric pulse.
How is the DNA taken up by bacteria during transformation?
The bound double-stranded DNA is nicked and cleaved into smaller fragments by membrane-bound endonucleases. The single strand of DNA then enters the bacterial cell through a membrane-spanning DNA translocation channel.
What is the role of recombination in bacterial transformation?
Recombination is the process by which the transformed DNA integrates into the bacterial chromosome, replacing a corresponding fragment. Recombination requires significant nucleotide sequence homology between the donor DNA and the bacterial chromosome.
What are some examples of bacteria that can undergo natural transformation?
Examples include Streptococcus pneumoniae, Haemophilus influenzae, Bacillus subtilis, and Neisseria species. These bacteria exhibit natural competence and have been extensively studied for their transformation abilities.
What are the applications of bacterial transformation?
Bacterial transformation has various applications in genetic engineering, biotechnology, antibiotic resistance studies, gene function and regulation studies, creation of genetically modified organisms (GMOs), development of molecular tools, and evolutionary and ecological studies.
How is the success of bacterial transformation determined in the laboratory?
In the laboratory, successful bacterial transformation can be identified by using selectable markers, such as antibiotic resistance genes carried on the transforming DNA. Only transformed cells carrying the marker gene will survive and grow on selective media.
Who discovered bacterial transformation?
The discovery of bacterial transformation is attributed to Frederick Griffith, an English bacteriologist, who made this significant observation in 1928. Griffith was studying Streptococcus pneumoniae, specifically different strains of the bacterium causing pneumonia. In his experiments, he observed that a non-virulent strain of the bacterium could become virulent and cause disease when it was in contact with heat-killed virulent strains of the same bacterium. This observation led to the concept of “transformation,” where genetic material from the heat-killed virulent strain was taken up by the non-virulent strain, resulting in a heritable change in its phenotype. Griffith’s discovery laid the foundation for further research on bacterial transformation and the understanding of genetic exchange mechanisms in bacteria.
- Griffith, F. (1928). The significance of pneumococcal types. Journal of Hygiene, 27(2), 113-159.
- Dubnau, D., & Blokesch, M. (2019). Mechanisms of DNA uptake by naturally competent bacteria. Annual Review of Genetics, 53, 217-237.
- Chen, I., & Dubnau, D. (2004). DNA uptake during bacterial transformation. Nature Reviews Microbiology, 2(3), 241-249.
- Johnsborg, O., & Håvarstein, L. S. (2009). Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiology Reviews, 33(3), 627-642.
- Claverys, J. P., & Martin, B. (2003). Bacterial “competence” genes: signatures of active transformation, or only remnants?. Trends in Microbiology, 11(4), 161-165.
- Chen, I., & Dubnau, D. (2003). DNA uptake during bacterial transformation. Methods in Enzymology, 358, 32-45.
- Redfield, R. J., Cameron, A. D., & Qian, Q. (2020). Heterologous transformation of Neisseria and other bacterial species: implications for genetic exchange. Trends in Microbiology, 28(5), 392-402.
- Dorer, M. S., & Fero, J. (2010). Bacterial transformation: the last step is the first. Microbe, 5(6), 280-286.
- Chen, I., Christie, P. J., & Dubnau, D. (2005). The ins and outs of DNA transfer in bacteria. Science, 310(5753), 1456-1460.
- Palchevskiy, V., & Finkel, S. E. (2009). Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. Journal of Bacteriology, 191(3), 601-611.
- Johnston, C., Martin, B., Fichant, G. et al. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12, 181–196 (2014). https://doi.org/10.1038/nrmicro3199
- Bacterial Transformation. (2023, February 18). West Hills College Lemoore. https://bio.libretexts.org/@go/page/79457
- https://www.thermofisher.com/in/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/molecular-cloning/transformation/bacterial-transformation-workflow.html
- frontiersin.org/articles/10.3389/fmicb.2017.02169/full
- https://www.pbslearningmedia.org/resource/biot11.sci.life.gen.transbact/transforming-bacteria/
- https://www.sigmaaldrich.com/IN/en/technical-documents/technical-article/genomics/advanced-gene-editing/transformation
- https://www.addgene.org/protocols/bacterial-transformation/
- https://www.khanacademy.org/science/biology/biotech-dna-technology/dna-cloning-tutorial/a/bacterial-transformation-selection