What is API (Analytical Profile Index) 20E Test?
The API (Analytical Profile Index) 20E test is a standardized biochemical identification system used for the identification of Enterobacteriaceae and other non-fastidious Gram-negative rods. It is developed as a miniaturized system where a number of biochemical reactions are carried out simultaneously in a single plastic strip. It is used commonly in clinical as well as industrial microbiology laboratories for routine identification of bacterial isolates.
It is the process in which twenty different biochemical tests are incorporated into small microtubes containing dehydrated substrates. These substrates are rehydrated by adding a bacterial suspension prepared from a pure culture. The system replaces the requirement of preparing and incubating multiple individual biochemical test media, thus saving time, reagents, and space in the laboratory. The reactions is standardized and gives reproducible results when performed under proper conditions.
In this test, a bacterial suspension is prepared and inoculated into all the microtubes of the API 20E strip. Some of the tests require anaerobic conditions and this is achieved by adding sterile mineral oil over the reaction cupules. The inoculated strip is incubated at 37°C for a period of 18–24 hours in a humid chamber. During incubation, the metabolic activity of bacteria leads to color changes either directly or after the addition of specific reagents.
After incubation, the reactions are read and interpreted based on color development. The tests are arranged in groups of three and each positive reaction is assigned a numerical value of 1, 2, or 4. These values are added together to form a seven-digit numerical profile. This profile number is then compared with a reference database or codebook to determine the identity of the bacterial species. The API 20E test is considered reliable, economical, and easy to perform, and therefore it is widely used for bacterial identification in microbiology laboratories.
Principle of API (Analytical Profile Index) Test for Bacteria
The principle of API (Analytical Profile Index) test is based on the ability of bacteria to produce specific biochemical reactions under controlled conditions. It is a standardized and miniaturized biochemical system where different metabolic properties of bacteria are tested simultaneously. It is mainly designed for identification of Enterobacteriaceae and other non-fastidious Gram-negative bacteria based on their enzymatic activities and substrate utilization pattern.
It is the process in which a plastic strip containing a series of microtubes (cupules) is used. Each microtube contains a dehydrated biochemical substrate. When a bacterial suspension prepared from a pure culture is added, these substrates are rehydrated. During incubation, the bacterial enzymes act on the substrates and metabolic reactions is carried out, leading to formation of acids, gases, or other end products.
The reactions occurring in the microtubes are detected by visible color changes. These color changes occur either due to pH indicators present in the medium or after the addition of specific reagents. Fermentation of carbohydrates produces acidic end products which changes the colour of the indicator, while some enzymatic reactions are detected only after adding reagents at the end of incubation.
The pattern of positive and negative reactions obtained after incubation represents the biochemical profile of the organism. These reactions are grouped and converted into a numerical code. This numerical profile is then compared with a standard database or codebook to determine the identity of the bacterial species. Thus, the API test works on the principle of biochemical differentiation and numerical identification of bacteria.
Objectives of API (Analytical Profile Index) 20E Test
The objectives of API (Analytical Profile Index) 20E test are as follows–
- To identify members of Enterobacteriaceae family based on their biochemical reactions.
- To identify other non-fastidious Gram-negative rod-shaped bacteria.
- To provide a standardized and miniaturized biochemical testing system in microbiology laboratories.
- To replace the use of multiple conventional biochemical test tubes with a single test strip.
- To obtain a characteristic biochemical profile of bacteria for species level identification.
- To assist in clinical diagnosis by accurate identification of pathogenic bacteria.
- To identify microorganisms in industrial and environmental samples such as food and water.
- To train students and laboratory personnel in biochemical identification techniques.
Requirements for API (Analytical Profile Index) 20E Test
The requirements for API (Analytical Profile Index) 20E test are as follows–
Bacterial specimen requirements
- Pure bacterial culture obtained from a single well isolated colony (18–24 hours old).
- The organism should belong to Enterobacteriaceae or other non-fastidious Gram-negative rods.
- Gram staining to confirm Gram-negative nature of bacteria.
- Oxidase test to be performed separately as it is required for final identification.
Kit components
- API 20E strip containing 20 microtubes with dehydrated biochemical substrates.
- Incubation tray with lid to maintain humid conditions.
- Result or reading sheet for recording biochemical reactions.
- Clip seal to store unused strips properly.
Reagents and media
- Sterile saline (0.85% NaCl) or API suspension medium for preparing bacterial suspension.
- Sterile mineral oil for creating anaerobic condition in specific tests.
- Ferric chloride reagent for TDA test.
- Kovac’s or James reagent for indole test.
- Voges–Proskauer reagents (VP1 and VP2).
- Nitrate reduction reagents (Nit 1, Nit 2 and zinc dust if required).
- Oxidase reagent for oxidase reaction.
Equipment and laboratory supplies
- Sterile Pasteur pipette or micropipette for inoculation.
- Distilled water to maintain humidity in incubation tray.
- McFarland standard (0.5) to adjust turbidity of bacterial suspension.
- Incubator maintained at 35–37°C.
- Marker pen for labeling the test tray.
- Bunsen burner to maintain aseptic conditions.
- API identification codebook or software for interpretation of numerical profile.
Procedure of API (Analytical Profile Index) Test
- Preparation –
- A pure culture of single organism is first isolated on suitable isolation media such as MacConkey agar and incubated for 18–24 hours.
- Oxidase test is performed separately from the colony as this result is required for the final identification profile.
- An incubation tray with lid is prepared and about 5 ml of distilled water is added into the honeycombed wells of the tray to create humid atmosphere and to prevent drying of strip.
- The strain reference number or patient ID is written on the elongated flap of the tray and not on the lid.
- Inoculum Preparation–
- A single well isolated colony is picked from the agar plate using sterile loop or pipette.
- The colony is emulsified in 5 ml of sterile 0.85% NaCl solution or API suspension medium.
- The suspension is mixed properly and turbidity is adjusted to match 0.5 McFarland standard.
- Inoculation of the Strip –
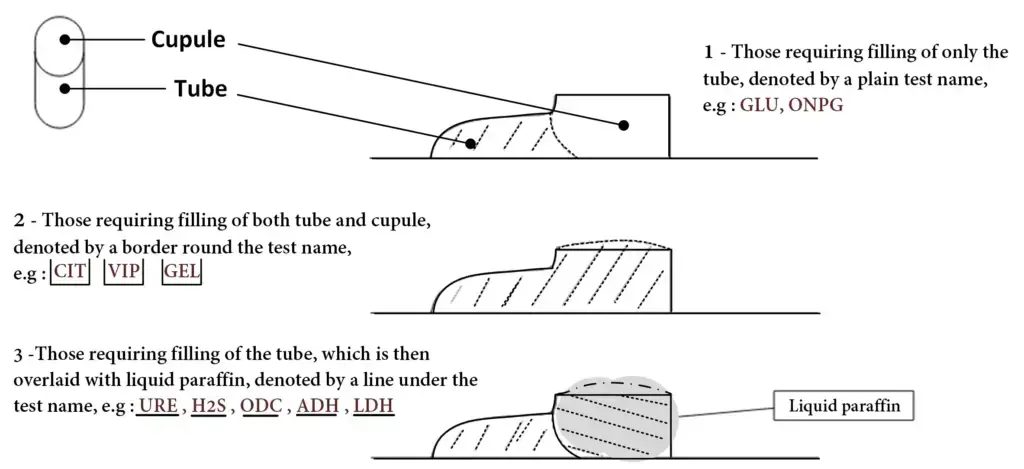
- The API 20E strip is placed inside the incubation tray and slightly tilted forward to avoid formation of air bubbles in the tubes.
- Using sterile Pasteur pipette the bacterial suspension is filled into the tubes of all 20 tests.
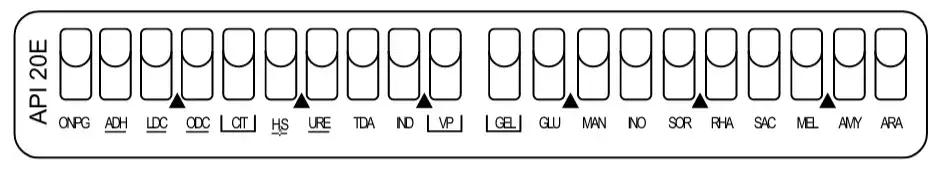
- For most of the tests only the tube portion is filled up to neck level leaving the cupule empty.
- For CIT, VP and GEL tests both tube and cupule is filled completely as these are aerobic tests.
- For ADH, LDC, ODC, H2S and URE tests the tube is filled with suspension and the cupule is filled with sterile mineral oil to create anaerobic condition.
- Incubation–
- The incubation tray is covered with lid carefully.
- The strip is incubated at 36°C ± 2°C or at 37°C for a period of 18 to 24 hours.
- Reading and Reagent Addition–
- After incubation the strip is read and spontaneous reactions are noted before adding reagents.
- If less than three reactions are positive the strip is re-incubated for additional 24 hours.
- One drop of TDA reagent is added to TDA cupule and reddish brown colour indicates positive result.
- One drop each of VP1 and VP2 reagent is added to VP cupule and results are read after 10 minutes.
- One drop of JAMES or Kovac’s reagent is added to IND test and formation of red colour or ring indicates positive reaction.
- Indole test is always performed at last as gaseous products may interfere with other tests.
- Interpretation–
- All test reactions are recorded as positive or negative using the reading chart.
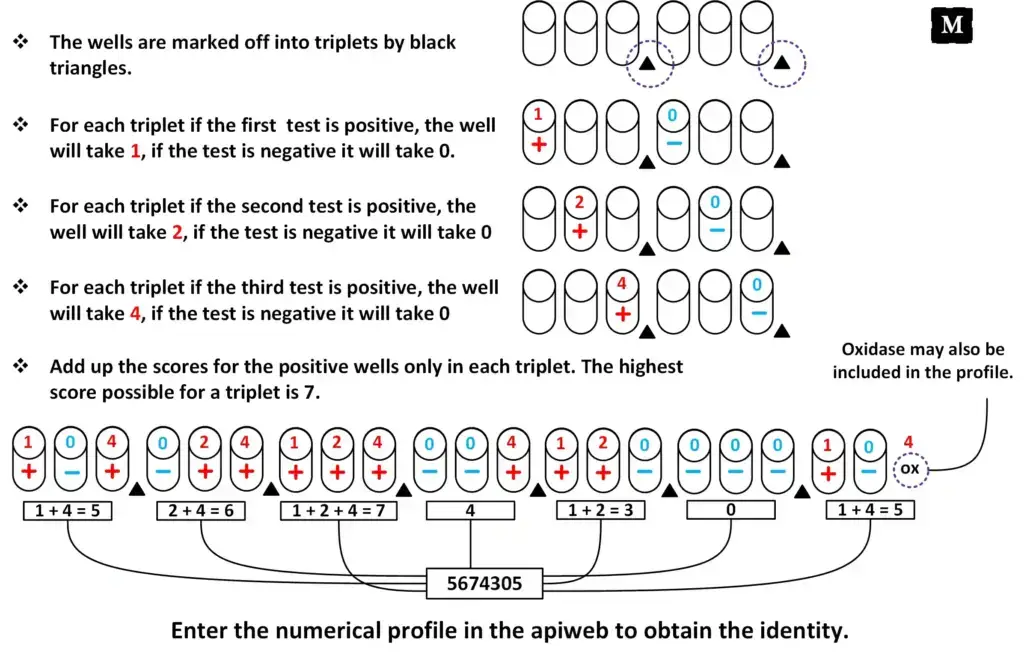
- The 20 biochemical tests are arranged into groups of three and values 1, 2 and 4 are assigned to positive reactions.
- The values of each group are added to obtain a seven digit numerical profile and oxidase result is also included.
- The obtained profile number is compared with API codebook or entered in identification software for final identification of organism.
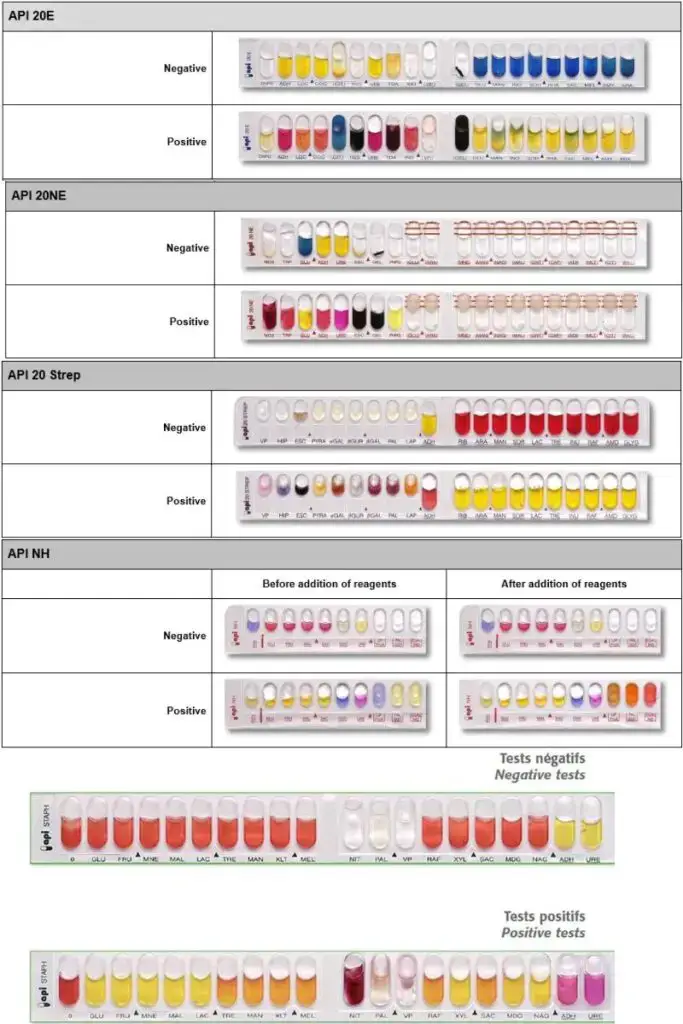
Result Interpretation of API 20E Test
The result interpretation of API 20E test is done on the basis of colour reactions and numerical profiling. The interpretation is carried out as follows–
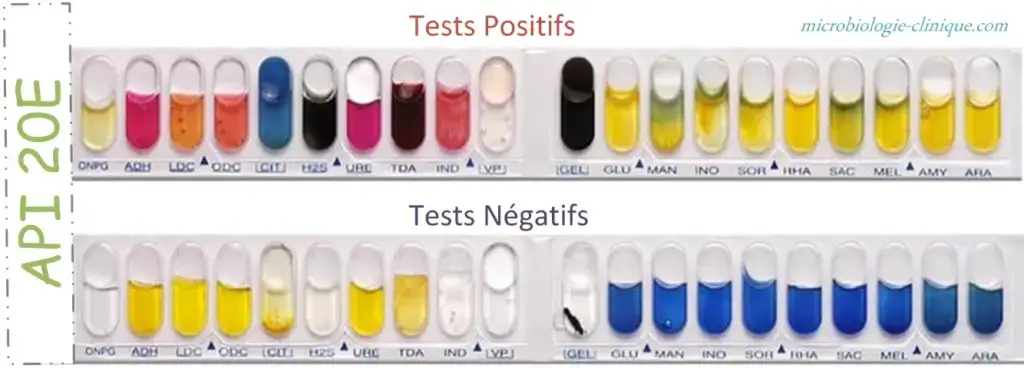
Observation of colour reactions
- Each microtube shows a colour change after incubation due to bacterial metabolic activity.
- The reactions are recorded as positive (+) or negative (–).
Carbohydrate fermentation tests
(GLU, MAN, INO, SOR, RHA, SAC, MEL, AMY, ARA)
- Yellow colour indicates positive reaction due to acid production.
- Blue or blue-green colour indicates negative reaction.
Amino acid decarboxylation and urease tests
(ADH, LDC, ODC, URE)
- Red or orange colour indicates positive reaction.
- Yellow colour indicates negative reaction.
Citrate utilisation test (CIT)
- Blue or blue-green colour indicates positive reaction.
- Pale green or yellow colour indicates negative reaction.
Hydrogen sulphide production (H2S)
- Black precipitate or thin black line indicates positive reaction.
- Absence of black colour indicates negative reaction.
Gelatin hydrolysis (GEL)
- Diffusion of black pigment indicates positive reaction.
- No diffusion indicates negative reaction.
β-galactosidase test (ONPG)
- Yellow colour indicates positive reaction.
- Colourless tube indicates negative reaction.
Reactions after addition of reagents
- TDA test shows reddish brown colour after adding ferric chloride for positive reaction.
- Indole test shows red ring after adding Kovac’s or James reagent for positive reaction.
- Voges–Proskauer test shows pink or red colour after adding VP reagents for positive reaction.
Numerical profile formation
- The twenty tests are arranged into groups of three.
- Positive reactions are assigned values of 1, 2 or 4 and negative reactions are given zero.
- The values of each group are added to form a single digit.
- A seven digit numerical profile is obtained including oxidase test result.
Final identification
- The numerical profile is compared with API codebook or identification software.
- The bacterial species is identified along with probability or confidence level.
Uses of API (Analytical Profile Index) 20E Test
The uses of API (Analytical Profile Index) 20E test are as follows–
- To identify and differentiate members of Enterobacteriaceae family.
- To identify other non-fastidious Gram-negative rod shaped bacteria.
- To assist in clinical diagnosis of bacterial infections.
- To identify causative agents of urinary tract infection, septicemia and other infections.
- To monitor food samples for pathogenic bacteria and spoilage organisms.
- To ensure quality control in pharmaceutical and industrial microbiology laboratories.
- To assess water quality by identification of coliform bacteria.
- To identify bacteria associated with veterinary infections.
- To support research work in bacterial taxonomy and classification.
- To teach biochemical identification techniques to students and laboratory trainees.
Advantages of API (Analytical Profile Index) 20E Test
The advantages of API (Analytical Profile Index) 20E test are as follows–
- It provides rapid identification of bacteria within 18–24 hours.
- It is economical as compared to automated identification systems.
- It is a standardized system and gives reproducible results in different laboratories.
- It reduces the need for preparing multiple conventional biochemical test media.
- It allows identification of bacteria up to species level.
- It is easy to perform and simple to interpret.
- It is a miniaturized system and requires less space and reagents.
- The test strips have long shelf life and are easy to store.
- It can identify Enterobacteriaceae as well as other non-fastidious Gram-negative rods.
- It is widely accepted as a reference method for bacterial identification.
Limitations of API (Analytical Profile Index) 20E Test
The limitations of API (Analytical Profile Index) 20E test are as follows–
- It is limited to identification of Enterobacteriaceae and other non-fastidious Gram-negative rods only.
- It cannot be used for Gram-positive bacteria or fastidious organisms.
- A pure culture is required and the test cannot be performed directly on clinical specimens.
- The incubation period is long and usually requires 18–24 hours.
- Re-incubation up to 48 hours may be needed when reactions are weak or insufficient.
- The test is highly dependent on correct inoculum density.
- Improper turbidity of bacterial suspension may give false positive or false negative results.
- Identification depends on availability of organism in the database.
- Some organisms such as Salmonella and Shigella require confirmatory serological tests.
- Environmental strains may give atypical or non-specific biochemical profiles.
- Technical errors like air bubble formation or improper filling of microtubes can affect results.
- Rare reversion of some reactions may cause difficulty in interpretation.
Download Protocols from API Galleries
| Galleries | Protocols |
|---|---|
| API 20E pdf | Download |
| API NE pdf | Download |
| API STREPT pdf | Download |
| API STAPH pdf | Download |
| API LISTERIA pdf | Download |
| API CANDIDA pdf | Download |
| API CH50 pdf | Download |
| API 20C pdf | Download |
FAQ
1. What is the API 20E test used for?
The API 20E test is used for identification of Enterobacteriaceae and other non-fastidious Gram-negative rods. It is commonly used in clinical, industrial and educational microbiology laboratories for routine bacterial identification.
2. What is the principle of the API 20E test?
The principle of API 20E test is based on biochemical activities of bacteria. It involves the reaction of bacterial enzymes with dehydrated substrates present in microtubes which produces colour changes. These reactions form a biochemical pattern that is used for identification.
3. How do you perform an API 20E test?
The test is performed by preparing a bacterial suspension from a pure culture and inoculating it into the microtubes of the API 20E strip. Some tests are overlaid with mineral oil and the strip is incubated at 35–37°C for 18–24 hours. After incubation, colour reactions are observed and reagents are added where required.
4. What organisms can be identified with the API 20E system?
The API 20E system can identify members of Enterobacteriaceae family and other non-fastidious Gram-negative rod shaped bacteria.
5. How do you interpret API 20E test results?
The results are interpreted by observing colour changes in each test and recording them as positive or negative. These results are converted into a numerical profile which is matched with a database or codebook for identification.
6. What are the individual tests included in the API 20E strip?
The strip includes carbohydrate fermentation tests, amino acid decarboxylation tests, enzyme activity tests, citrate utilization, hydrogen sulphide production, gelatin hydrolysis and β-galactosidase test.
7. What reagents are needed for the API 20E test?
Reagents such as ferric chloride for TDA test, Kovac’s or James reagent for indole test, Voges–Proskauer reagents, nitrate reagents and oxidase reagent are required for interpretation.
8. How is a profile number determined in the API 20E test?
The profile number is determined by grouping the tests into triplets and assigning values of 1, 2 or 4 to positive reactions. The values are added to obtain a seven digit numerical code.
9. What is the incubation time for the API 20E test?
The incubation time for API 20E test is usually 18–24 hours. In some cases re-incubation up to 48 hours may be required.
10. What is the API 20E test kit composed of?
The kit is composed of API 20E strips with dehydrated substrates, incubation tray with lid, result sheet and storage accessories.
11. What is the purpose of adding mineral oil in the API 20E test?
Mineral oil is added to create anaerobic conditions for certain biochemical reactions such as decarboxylation and urease tests.
12. What is the significance of the oxidase test in conjunction with API 20E?
The oxidase test helps in differentiating Enterobacteriaceae from other Gram-negative bacteria and is used as an additional test for final identification.
13. Can the API 20E test identify Gram-positive bacteria?
No, the API 20E test cannot identify Gram-positive bacteria. It is designed only for Gram-negative rods.
14. Where can I find the API 20E codebook or identification software?
The API 20E codebook is provided with the kit and identification software such as APIWEB is available through the manufacturer.
15. What are the advantages of the API 20E system?
The API 20E system is rapid, economical, standardized and easy to use. It requires less space and reagents and provides reliable identification up to species level.
- Acharya, T. (2024, February 28). API 20E Test System: Results and Interpretations. Microbe Online.
- American Society for Microbiology. (2012, November 1). Gelatin Hydrolysis Test Protocol.
- Aryal, S. (2022, August 10). API (Analytical Profile Index) 20E Test – Procedure, Uses and Interpretation. Microbiology Info.
- Aryal, S. (2022, August 10). Nitrate Reduction Test – Principle, Procedure, Uses and Interpretation. Microbiology Info.
- bioMérieux. (2002, October). API 20 E: Identification system for Enterobacteriaceae and other non-fastidious Gram-negative rods [Package insert].
- bioMérieux. (2003, October). API 20 NE: Identification system for non-fastidious, non-enteric Gram-negative rods [Package insert].
- bioMérieux. (2019, June). API 20 E [Package insert].
- bioMérieux. (n.d.). API® ID Strip Range and APIWEB™.
- bioMérieux. (n.d.). API® ID STRIPS.
- bioMérieux. (n.d.). Rapid 20 E: System for the identification of Enterobacteriaceae in 4 hours [Package insert].
- bioMérieux. (n.d.). Reference Guide: API® & ID 32 Identification databases.
- Chegg. (n.d.). Exercise 8: Use of the API 20E System to Identify Gram Negative Bacteria.
- Lindquist, J. (2010, June 1). Bacterial Identification – The API-20E System.
- Martinez-Urtaza, J., Lozano-Leon, A., Viña-Feas, A., de Novoa, J., & Garcia-Martin, O. (2006). Differences in the API 20E biochemical patterns of clinical and environmental Vibrio parahaemolyticus isolates. FEMS Microbiology Letters, 255(1), 75–81.
- Montgomery County Community College. (n.d.). Title: API 20E Microbial Identification SOP. Biomanufacturing.org.
- O’Hara, C. M., Rhoden, D. L., & Miller, J. M. (1992). Reevaluation of the API 20E identification system versus conventional biochemicals for identification of members of the family Enterobacteriaceae: a new look at an old product. Journal of Clinical Microbiology, 30(1), 123–125.
- Reynolds, J. (2021, August 1). 43: API-20E multitest strip. Biology LibreTexts.
- Sigma-Aldrich. (2018). 73426 Nitrate Reduction Test [Technical Datasheet].
- Trotman, D. [Diann Trotman]. (n.d.). API 20E Part 1: Setup [Video]. YouTube.
- Wikipedia. (n.d.). Analytical profile index.
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.