What is Prokaryotic DNA Replication?
- Prokaryotic DNA replication is a fundamental biological process where prokaryotes, such as bacteria, duplicate their DNA to ensure genetic continuity during cell division. This intricate process ensures that each daughter cell receives an accurate copy of the genetic material, crucial for their survival and function.
- DNA replication in prokaryotes follows a semi-conservative method. This means each newly formed DNA molecule consists of one original parental strand and one newly synthesized daughter strand. This method helps preserve the genetic information across generations.
- Replication in prokaryotes, particularly well-studied in Escherichia coli (E. coli), begins at a unique origin of replication known as OriC. Here, replication initiates bi-directionally, meaning it proceeds in two opposite directions from the origin, facilitating efficient copying of the entire DNA molecule.
- The process of prokaryotic DNA replication can be broken down into three main steps: initiation, elongation, and termination.
- During initiation, several proteins, including initiator proteins, bind to the OriC region, unwinding the DNA to create replication forks. These forks are critical points where the DNA strands are separated, allowing replication to begin.
- Next, in the elongation phase, DNA polymerase enzymes add nucleotides to the growing DNA strand, complementary to the template strand. This phase involves a complex interaction of various proteins and enzymes, ensuring the accurate and rapid synthesis of the new DNA strands.
- Finally, termination occurs when the replication forks meet, completing the replication process. Specific termination sequences signal the end of replication, ensuring that the entire genome has been accurately copied.
- Prokaryotic DNA replication is an efficient and highly regulated process, critical for the survival and proliferation of prokaryotic organisms. Understanding this process in detail provides valuable insights into the fundamental mechanisms of life and can inform various applications in biotechnology and medicine.
Structure of Ori C
OriC (Origin of Chromosome) is the specific locus in Escherichia coli where DNA replication is initiated. This critical region is located at approximately the 84.5 mpu position on the E. coli genome, situated opposite to the replication termination site. The structure of OriC is characterized by two key regions: the 9-mer and the 13-mer.
- 9-mer Region (DNA-A Box):
- Structure: The 9-mer is a conserved sequence of nine nucleotides found within OriC. This region is also referred to as the DNA-A box.
- Function: The 9-mer serves as the primary binding site for the DnaA protein, a key initiator of DNA replication. DnaA binds specifically to this sequence, facilitating the formation of the DnaA-oriC complex.
- Role in Replication Initiation: Upon binding to the 9-mer, DnaA promotes the formation of a multi-protein complex that is essential for unwinding the DNA helix and recruiting other replication factors.
- 13-mer Region:
- Structure: The 13-mer consists of a repeating sequence of thirteen nucleotides characterized by a high proportion of adenine-thymine (A-T) base pairs.
- Function: This region is known for its lower melting temperature compared to regions with higher guanine-cytosine (G-C) content.
- Role in Replication Initiation: The high A-T content of the 13-mer facilitates the separation of the DNA strands. This is crucial for the formation of the replication bubble, allowing access for the replication machinery to initiate DNA synthesis.
Mechanism of prokaryotic dna replication
The synthesis of a DNA molecule can be divided into three stages:
- Initiation
- Elongation
- Termination
distinguished both by the reactions taking place and by the enzymes required. As you will find here and in the next two chapters, synthesis of the major information containing biological polymers-DNAs, RNAs, and proteins-can be understood in terms of these same three stages, with the stages of each pathway having unique characteristics. The events described below reflect information derived primarily from in vitro experiments using purified E col’i proteins, although the principles are highly conserved in all replication systems.
Enzymes involved in prokaryotic dna replication
The enzymes involved in DNA replication are vital for ensuring the accurate duplication of the genetic material. These enzymes each play a specific role, coordinating to achieve efficient and precise DNA synthesis. Below is a detailed examination of the key enzymes and their functions during DNA replication.
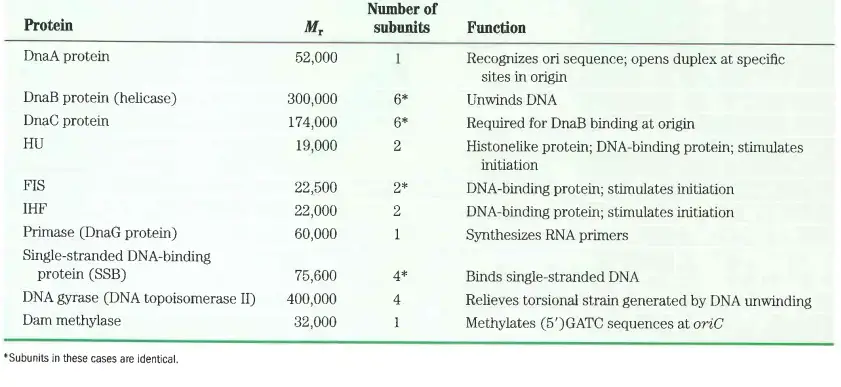
- Helicases
- Function: Unwind the DNA helix at the start of replication.
- Mechanism: These enzymes break the hydrogen bonds between the DNA strands, creating two single strands that serve as templates for replication.
- Single-Strand Binding (SSB) Proteins
- Function: Bind to the single strands of unwound DNA.
- Role: Prevent reformation of the DNA helix during replication, stabilizing the single-stranded DNA and protecting it from degradation.
- Primase
- Function: Synthesizes RNA primers.
- Initiation: These RNA primers are necessary for the initiation of DNA chain synthesis, providing a starting point for DNA polymerases to begin DNA synthesis.
- DNA Polymerase III (DNAP III)
- Function: Elongates the DNA strand.
- Specifics: Adds deoxyribonucleotides to the 3′ end of the chain.
- Directionality: Synthesis occurs in the 5′ to 3′ direction, which is a fundamental characteristic of DNAP III’s function.
- DNA Polymerase I (DNAP I)
- Function: Replaces RNA primers with DNA.
- Process: Removes RNA primers and fills in the gaps with the appropriate deoxynucleotides, ensuring the continuity of the DNA strand.
- DNA Topoisomerase I
- Function: Relaxes the DNA helix during replication.
- Action: Creates a temporary nick in one of the DNA strands to alleviate the tension generated by the unwinding of the helix.
- DNA Topoisomerase II
- Function: Relieves the strain on the DNA helix.
- Mechanism: Introduces supercoils into the DNA helix by creating nicks in both strands, thereby preventing overwinding and tangling.
- DNA Ligase
- Function: Forms a 3′-5′ phosphodiester bond.
- Role: Connects adjacent fragments of DNA, such as Okazaki fragments on the lagging strand, ensuring the integrity and continuity of the newly synthesized DNA.
| Enzyme | Function | Details |
|---|---|---|
| Helicases | Unwind the DNA helix | Break hydrogen bonds between DNA strands, creating single strands for replication. |
| SSB Proteins | Bind to single strands of unwound DNA | Stabilize single-stranded DNA and prevent reformation of the DNA helix during replication. |
| Primase | Synthesizes RNA primers | Provides starting points for DNA polymerases to initiate DNA synthesis. |
| DNA Polymerase III | Elongates DNA strand | Adds deoxyribonucleotides to the 3′ end of the chain; synthesizes in the 5′ to 3′ direction. |
| DNA Polymerase I | Replaces RNA primers with DNA | Removes RNA primers and fills in gaps with deoxynucleotides. |
| DNA Topoisomerase I | Relaxes the DNA helix | Creates temporary nicks in one DNA strand to alleviate tension from unwinding. |
| DNA Topoisomerase II | Relieves strain on the DNA helix | Introduces supercoils by creating nicks in both DNA strands to prevent overwinding and tangling. |
| DNA Ligase | Forms 3′-5′ phosphodiester bonds between DNA fragments | Connects adjacent DNA fragments, ensuring the continuity and integrity of the newly synthesized DNA. |
Steps of DNA Replication
Here is the detailed explanation of the steps involved in DNA replication:
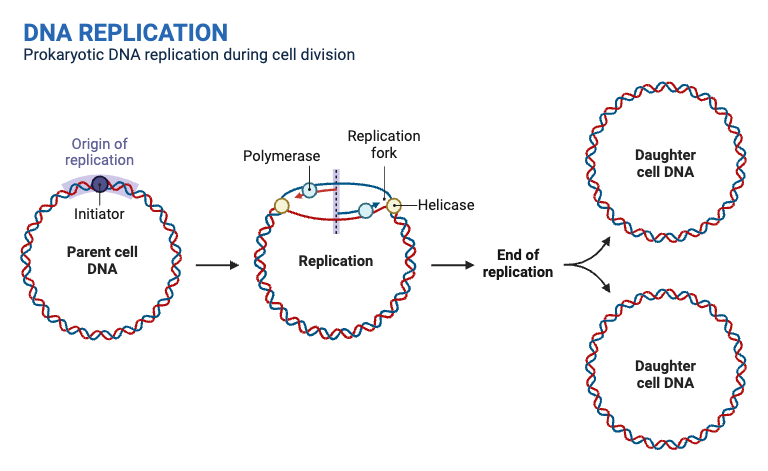
- Initiation at the Origin of Replication
- Origin of Replication: DNA replication begins at a specific sequence known as the origin of replication.
- Unwinding of Double Helix: Enzymes such as helicase unwind the DNA double helix, creating accessible single strands for replication.
- Formation of Replication Forks
- Replication Forks: Helicase activity generates a pair of replication forks at the origin, where the DNA is actively unwound.
- Stabilization: Single-Strand Binding (SSB) proteins stabilize the unwound DNA, preventing it from re-forming the double helix. DNA topoisomerases alleviate the tension in the DNA helix by creating temporary nicks.
- Synthesis of RNA Primers
- Primase Activity: Primase synthesizes short RNA primers (approximately 10 bases long) at the replication forks.
- Primer Role: These RNA primers provide the starting points for DNA polymerase to begin DNA synthesis.
- Elongation of the Leading Strand
- Continuous Synthesis: DNA Polymerase III (DNAP III) elongates the leading strand continuously in the 5′ to 3′ direction, following the replication fork.
- Elongation of the Lagging Strand
- Discontinuous Synthesis: The lagging strand is synthesized discontinuously, also in the 5′ to 3′ direction, through the formation of short DNA segments known as Okazaki fragments.
- Fragment Synthesis: Multiple RNA primers are laid down by primase, allowing DNAP III to synthesize Okazaki fragments.
- Primer Removal and Replacement
- DNA Polymerase I Activity: DNAP I removes RNA primers from both the leading and lagging strands.
- Gap Filling: DNAP I fills in the resulting gaps with the appropriate deoxynucleotides, ensuring continuity of the DNA strand.
- Sealing of DNA Fragments
- DNA Ligase Function: DNA ligase seals the nicks between Okazaki fragments and the gaps left after primer removal.
- Formation of Continuous Strands: This sealing forms continuous and complete DNA strands, finalizing the replication process.
A. Initiation of Prokaryotic DNA Replication
Protein required for initiation of Replication in Prokaryotes
Step 1: Oric
The origin of replication in E. coli, known as oriC, is a critical region of the bacterial chromosome where DNA replication begins. It spans 245 base pairs and contains specific DNA sequences essential for initiating replication. The structure and function of oriC are well conserved among various bacterial species, reflecting its fundamental role in the replication process. Below is a detailed examination of the components and functions of oriC.
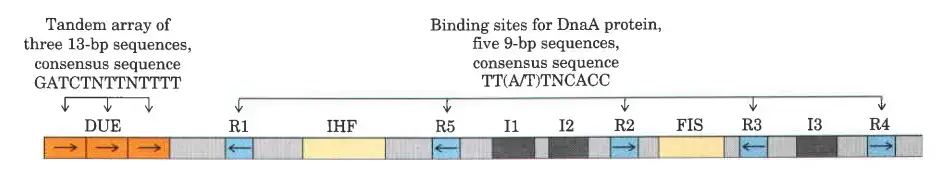
- Structure of oriC
- Length and Composition: oriC is 245 base pairs long, comprising several specific sequence motifs necessary for the initiation of DNA replication.
- Key Sequence Elements
- R Sites:
- Definition: Five repeats of a 9-base pair sequence.
- Function: Serve as binding sites for DnaA, the key initiator protein. DnaA binding is crucial for the unwinding of the DNA at the origin.
- DNA Unwinding Element (DUE):
- Composition: A region rich in adenine-thymine (A) base pairs.
- Function: The Aregion facilitates the unwinding of the DNA helix, as Abase pairs are less stable than guanine-cytosine (G) pairs.
- R Sites:
- Binding Sites for Additional Proteins
- I Sites:
- Function: Specific binding sites for DnaA proteins, enhancing the binding affinity and stability of the initiation complex.
- IHF (Integration Host Factor):
- Role: A DNA-binding protein that bends the DNA, facilitating the assembly of the initiation complex.
- FIS (Factor for Inversion Stimulation):
- Role: Another DNA-binding protein that aids in the proper formation of the initiation complex.
- HU Protein:
- Function: A histone-like bacterial protein that stabilizes the open complex formed during the initiation of replication. Although it does not have a specific binding site, HU plays a significant role in maintaining the structure necessary for replication.
- I Sites:
- Functional Overview
- DnaA Binding: DnaA proteins bind to the R and I sites, causing the DNA to bend and partially unwind at the DUE.
- Assembly of Initiation Complex: IHF and FIS proteins further facilitate the bending and stabilization of the DNA, promoting the formation of the open complex necessary for replication.
- DNA Unwinding: The ADUE region is particularly susceptible to unwinding, allowing helicase and other replication machinery to access and replicate the DNA strands.
Step 2: Oric Recognize by DnaA
The initiation of DNA replication in E. coli is a complex process involving multiple proteins and enzymes. Central to this process is the DnaA protein, a member of the AAA+ ATPase family, which plays a critical role in recognizing the origin of replication, oriC. Below is a detailed explanation of the steps involved in this recognition process.
- Formation of the Pre-replication Complex
- Protein Assembly:
- At the onset of replication, at least 10 different enzymes and proteins assemble to open the DNA helix.
- This assembly sets up a complex required for subsequent replication steps.
- Protein Assembly:
- Role of DnaA Protein
- AAA+ ATPase Family:
- DnaA is part of the AAA+ ATPase family, known for its role in various cellular activities.
- It forms oligomers and hydrolyzes ATP at a slow rate, acting as a molecular switch.
- Active vs. Inactive Forms:
- The ATP-bound form of DnaA is active, while the ADP-bound form is inactive.
- This ATP hydrolysis enables the protein to transition between active and inactive states.
- AAA+ ATPase Family:
- Binding to oriC
- Formation of DnaA Helix:
- Eight ATP-bound DnaA molecules oligomerize to form a right-handed helical structure.
- This structure wraps around the R and I sites in oriC.
- Differential Binding:
- DnaA has a higher affinity for R sites than I sites.
- It binds to R sites equally well in both ATP and ADP-bound forms.
- I sites bind exclusively to the ATP-bound form, distinguishing active DnaA.
- Formation of DnaA Helix:
- Induction of Supercoiling
- DNA Wrapping:
- The DNA wraps tightly around the DnaA-oriC complex, creating a positive supercoil.
- Effect on DUE Region:
- The strain from the supercoiling destabilizes the adjacent ADNA unwinding element (DUE) region.
- This destabilization is critical for initiating DNA unwinding.
- DNA Wrapping:
- Involvement of Additional Proteins
- DNA-Binding Proteins:
- The complex at oriC also includes Hu, IHF (Integration Host Factor), and FIS (Factor for Inversion Stimulation).
- These proteins facilitate DNA bending, further promoting the formation of the replication initiation complex.
- DNA-Binding Proteins:
Therefore, the recognition of oriC by DnaA involves a precise and coordinated sequence of events. The assembly of the pre-replication complex, the ATP-dependent activation of DnaA, and the differential binding to R and I sites are crucial steps. The induced supercoiling and the involvement of additional DNA-binding proteins ensure that the DNA unwinding element is properly destabilized, setting the stage for the initiation of DNA replication. This intricate process underscores the complexity and efficiency of bacterial DNA replication mechanisms.
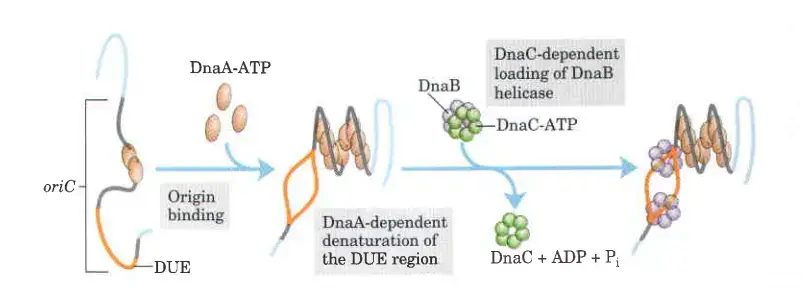
Step 3: DnaC Protein
In the intricate process of DNA replication, the DnaC protein plays a crucial role, particularly in the loading of the DnaB helicase onto the DNA strands. This step ensures that the replication machinery is properly assembled and ready for the synthesis of new DNA strands. Below is a detailed and sequential explanation of how DnaC facilitates this process.
- Binding of DnaC to DnaB
- AAA+ ATPase Function:
- Similar to DnaA, DnaC is a member of the AAA+ ATPase family.
- Its primary role is to bind to the DnaB helicase and facilitate its attachment to the DNA strands.
- AAA+ ATPase Function:
- Formation of the DnaB-DnaC Complex
- Ring-Shaped DnaB Helicase:
- The DnaB protein forms a ring-shaped helicase structure.
- This helicase forms a tight complex with six DnaC subunits, each bound to ATP.
- Opening of the DnaB Ring:
- The interaction between DnaC and DnaB opens the DnaB ring, allowing it to encircle the DNA strands.
- This process is further aided by an interaction between DnaB and the DnaA proteins already bound at the origin of replication.
- Ring-Shaped DnaB Helicase:
- Loading of DnaB onto DNA Strands
- Placement on DNA Strands:
- One of the ring-shaped DnaB hexamers is loaded onto each of the single DNA strands within the denatured DNA unwinding element (DUE) region.
- This ensures that each DNA strand is primed for replication.
- Placement on DNA Strands:
- Hydrolysis of ATP
- ATP Hydrolysis and Release of DnaC:
- The ATP bound to the DnaC subunits is hydrolyzed, a reaction catalyzed by the presence of water.
- This hydrolysis releases the DnaC protein from the complex, but the DnaB helicase remains tightly bound to the DNA.
- ATP Hydrolysis and Release of DnaC:
Summary of Functions and Processes
- DnaC Binding to DnaB:
- Facilitates the interaction between DnaB and the DNA strands.
- DnaB-DnaC Complex Formation:
- Ensures the correct assembly of the helicase structure.
- Loading DnaB onto DNA:
- Positions the helicase for effective unwinding of the DNA helix.
- ATP Hydrolysis:
- Releases DnaC, leaving DnaB bound and functional on the DNA strands.
Therefore, DnaC acts as a crucial mediator in the assembly of the replication machinery, specifically by loading the DnaB helicase onto the DNA. This step is essential for the continuation of the DNA replication process, ensuring that the DNA strands are properly unwound and accessible for the synthesis of new strands. The precise interaction and regulation by ATP hydrolysis underline the complex yet highly coordinated nature of DNA replication.
Step 4: Role of DnaB Protein, SSB, and DNA Gyrase in DNA Replication
The loading of the DnaB helicase is a pivotal step in the initiation of DNA replication. This step sets the stage for the unwinding of the DNA helix and the establishment of replication forks. Here is a detailed and sequential explanation of how DnaB, single-stranded DNA-binding proteins (SSB), and DNA gyrase function in this process.
- DnaB Protein: The Replicative Helicase
- Unwinding DNA:
- DnaB helicase is essential for unwinding the DNA double helix.
- It moves along the single-stranded DNA in a 5′-to-3′ direction.
- The two DnaB helicases bound to the DNA strands move in opposite directions, creating two replication forks.
- Interaction with Other Proteins:
- DnaB is the central protein to which other replication fork proteins are connected.
- It interacts with various components, including the τ subunits of the DNA polymerase III holoenzyme, facilitating the polymerase’s function.
- Unwinding DNA:
- Single-Stranded DNA-Binding Protein (SSB)
- Stabilizing Separated Strands:
- As the DNA strands are separated at the replication fork, multiple SSB molecules bind to the single-stranded DNA.
- This binding stabilizes the separated strands, preventing them from reannealing or forming secondary structures.
- Protection and Prevention:
- SSBs protect the single-stranded DNA from nucleases.
- They also prevent the formation of hairpin loops, which can hinder the replication process.
- Stabilizing Separated Strands:
- DNA Gyrase (DNA Topoisomerase II)
- Relieving Topological Stress:
- DNA gyrase, a type of topoisomerase, alleviates the topological stress generated by the unwinding of the DNA helix.
- It introduces negative supercoils into the DNA, counteracting the positive supercoils formed ahead of the replication fork.
- Maintaining DNA Integrity:
- By relieving the torsional strain, DNA gyrase ensures that the DNA remains intact and accessible for replication machinery.
- Relieving Topological Stress:
Summary of Functions and Processes
- DnaB Protein:
- Central Role: Unwinds the DNA double helix and facilitates the formation of replication forks.
- Protein Interactions: Connects with other replication proteins to coordinate the replication process.
- Single-Stranded DNA-Binding Protein (SSB):
- Stabilization: Binds to single-stranded DNA to stabilize and protect it.
- Prevention: Prevents reannealing and secondary structure formation.
- DNA Gyrase (DNA Topoisomerase II):
- Stress Relief: Introduces negative supercoils to relieve topological stress.
- Integrity Maintenance: Ensures DNA remains intact and accessible for replication.
Regulation of DNA Replication Initiation
- Control Mechanism:
- The initiation of DNA replication is the only known regulated step, ensuring that replication occurs only once per cell cycle.
- Although the exact regulatory mechanisms are not fully understood, genetic and biochemical studies have provided insights into several regulatory pathways.
Step 5: Protein Hda
- Once DNA polymerase III and the B subunits have been loaded onto the DNA, which marks the end of the initiation phase, the protein Hda binds to the B subunits and interacts with DnaA to speed up the hydrolysis of the ATP that is bound to DnaA.
- Hda is also a AAA+ ATPase that is very similar to DnaA. (its name is derived from homologous to DnaA).
- This breakdown of ATP causes the DnaA complex at the beginning of the cell to fall apart.
- The protein changes between its inactive (with bound ADP) and active (with bound ATP) forms every 20 to 40 minutes. This happens because DnaA slowly lets go of ADP and rebinds ATP.
Step 6: DNA adenine methylation
- Methylation of DNA and interactions with the bacterial plasma membrane can change when replication starts.
- The Dam methylase methylates the N6 spot of adenine in the palindromic sequence (5′)GATC, which is part of the ori,C DNA. (DNA adenine methylation is what “Dam” means in biochemistry.)
- The E. coli ori,C region has a lot of GATC sequences. It has 11 of them in 245 bp, while the average number of GATC sequences in the whole E. coli chromosome is 1 in 256 bp.
- The DNA is hemimethylated right after replication. The parent strands have methylated ori,C sequences, but the new strands don’t. The protein SeqA now binds to the hemimethylated oriC sequences and keeps them from moving. This is done by the interaction with the plasma membrane (how this happens is not known).
- After some time, ori,C is released from the plasma membrane, SeqA breaks apart, and the DNA must be fully methylated by Dam methylase before it can bind to DnaA again and start a new round of replication.
B. Elongation of Prokaryotic DNA Replication
- During the elongation phase of replication, Ieading strand synthesis and Lagging strand synthesis take place. These are two separate but related processes.
- At the replication fork, there are several enzymes that are important for making both strands. First, DNA helicases unwind the parent DNA. Then, topoisomerases relieve the topological stress that is caused.
- Then, SSB is used to keep each strand in place. Synthesis of the leading and lagging strands is very different from this point on.
Leading strand synthesis
- The simpler of the two processes, leading strand synthesis starts with primase (DnaG protein) making a short (10–60 nucleotide) RNA primer at the replication origin.
- DnaG and DnaB helicase work together to make this reaction happen. The primer is made in the opposite direction that the DnaB helicase is moving.
- In reality, the DnaB helicase moves along the strand that becomes the lagging strand during DNA synthesis. However, the first primer that is laid down during the first DnaGDnaB interaction starts DNA synthesis on the leading strand in the opposite direction.
- A DNA polymerase III complex linked to the DnaB helicase on the other DNA strand adds deoxyribonucleotides to this primer.
- Then, the process of making the leading strand keeps going at the same rate as the unwinding of DNA at the replication fork.
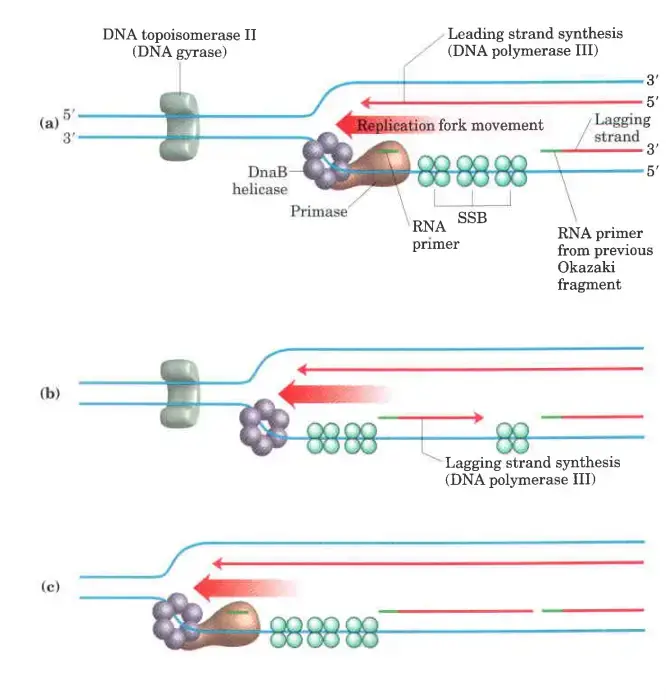
Lagging strand synthesis
- We’ve already said that lagging strand sythesis is done in short Okazaki fragments.
- First, primase makes an RNA primer. Then, just like in leading strand synthesis, DNA polymerase III binds to the RNA primer and adds deoxyribonucleotides.
- On this level, putting together each Okazaki fragment seems easy, but it’s actually quite hard. The coordination of leading and lagging strand synthesis is what makes it hard.
- A single asymmetric DNA polymerase III dimer makes both strands. This is done by looping the DNA of the Lagging strand, which brings the two points of polymerization together.
Synthesis of Okazaki fragment
The synthesis of Okazaki fragments is a crucial aspect of DNA replication on the lagging strand. This process involves a complex series of enzymatic activities, orchestrated with precision to ensure accurate DNA duplication.
- Formation of Primosomes:
- Primosomes are key functional units within the replication complex. They consist of two essential components: DnaB helicase and DnaG primase. DnaB helicase is responsible for unwinding the DNA double helix at the replication fork, while DnaG primase synthesizes short RNA primers necessary for DNA polymerase activity.
- Role of DNA Polymerase III:
- DNA Polymerase III plays a central role in DNA replication. Its core polymerase subunits are continuously engaged in synthesizing the leading strand. On the lagging strand, however, the core polymerase subunits cycle from one Okazaki fragment to the next. This movement is facilitated by the formation of a looped structure in the lagging strand template.
- Unwinding and Primer Synthesis:
- As DNA Polymerase III progresses along the lagging strand in the 5′ to 3′ direction, DnaB helicase unwinds the DNA in front of it. In certain instances, DnaG primase collaborates with DnaB helicase to generate a short RNA primer. This primer serves as the starting point for DNA synthesis.
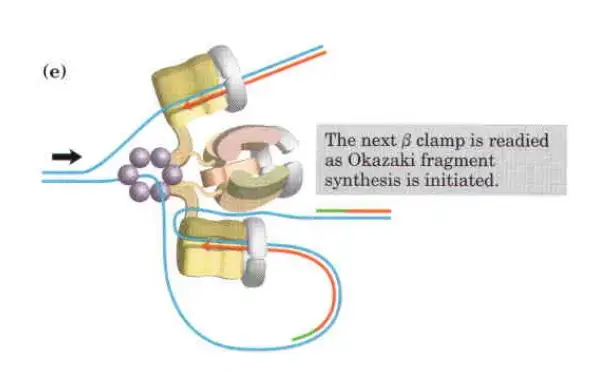
- Clamp Loading and Synthesis:
- Following primer synthesis, the clamp loading complex of DNA Polymerase III deposits a new β sliding clamp onto the primer. This clamp is essential for the stability and processivity of DNA polymerase during the elongation phase.
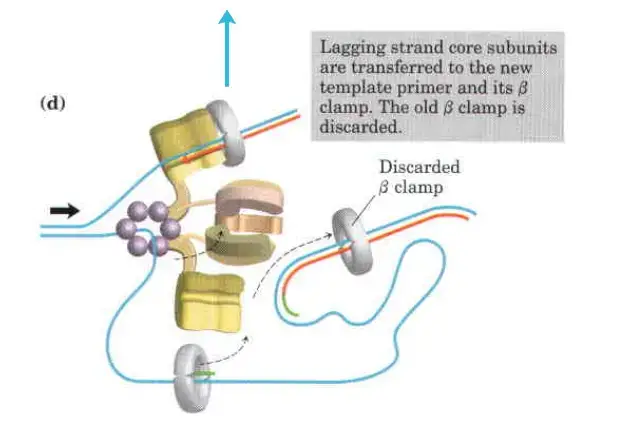
- Detachment and Re-association:
- Upon completing an Okazaki fragment, the core subunits of DNA Polymerase III detach from the β sliding clamp and the newly synthesized fragment. These subunits then re-associate with a new β sliding clamp to initiate the synthesis of the next Okazaki fragment.
- Function of the Replisome:
- The replisome is a complex of proteins located at the replication fork, playing a critical role in coordinating DNA synthesis. It integrates the activities of various enzymes, ensuring the efficient and accurate replication of DNA.
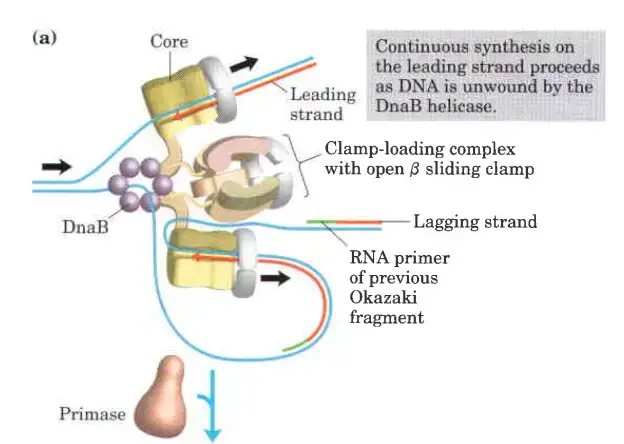
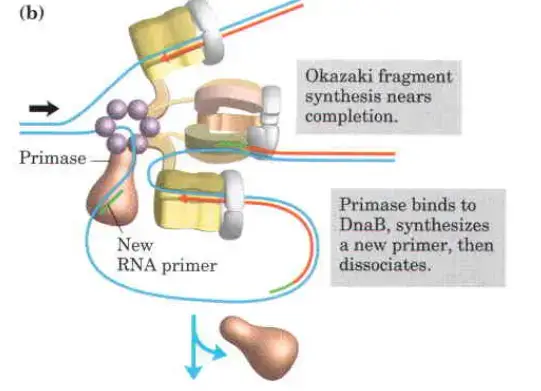
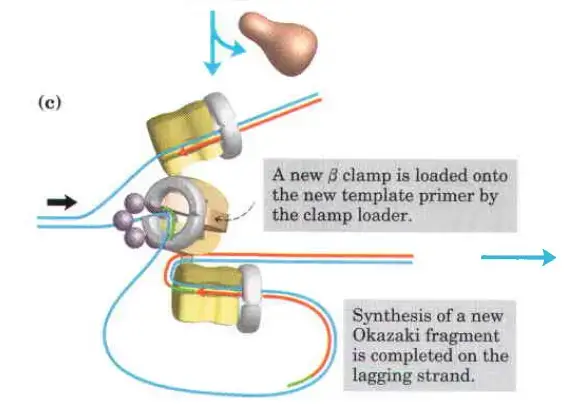
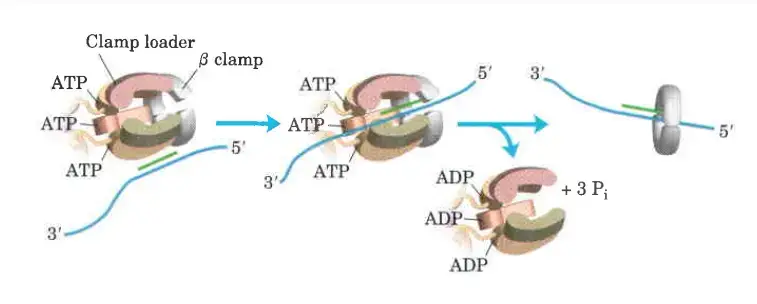
The clamp Loading complex of DNA polymerase III
- The clamp Loading complex of DNA polymerase III, consisting of parts of the two τ subunits along with the γ, δ, and δ’ subunits, is also an AAA+ ATPase.
- Together, ATP and the novel B sliding clamp form a stable combination. The dimeric clamp undergoes strain as a result of the binding, and the ring opens up at one of the subunit interfaces.
- Through this rip, the newly primed lagging strand is introduced to the ring.
- Hydrolysis of ATP by the clamp loader then allows the B sliding clamp to close around the DNA.
DNA synthesis
- The replisome is responsible for the rapid synthesis of DNA, at a rate of -1,000 nucleotides per second per strand (leading and lagging).
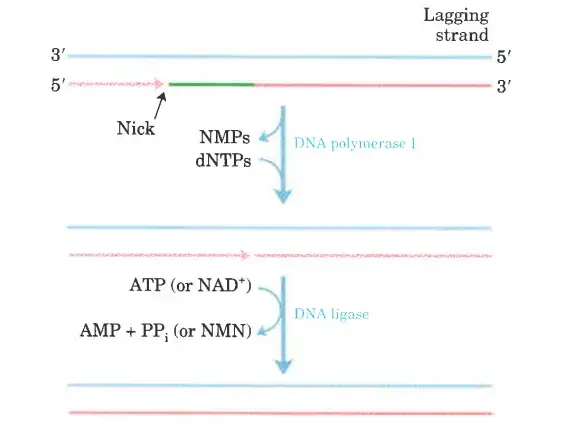
- DNA ligase seals the residual nick in an Okazaki fragment after DNA polymerase I has removed the RNA primer.
- DNA ligase is an enzyme that catalyses the creation of a phosphodiester link between a 3′ hydroxyl and a 5′ phosphate on opposite ends of DNA strands.
- Adenylylation is required to activate the phosphate. The ATP is used by DNA ligases that have been purified from viruses and eukaryotes.
- Bacterial DNA ligases are distinct in that many of them derive the AMP activating group from the coenzyme NAD+, which typically catalyses hydride transfer events. DNA ligase is another DNA metabolic enzyme that has emerged as a crucial tool in recombinant DNA research.
Proteins required for replisome
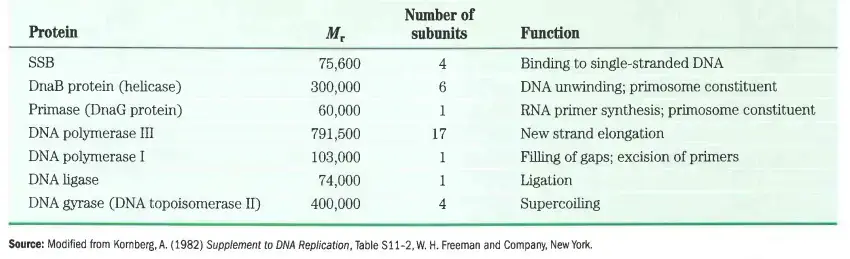
C. Termination of Prokaryotic DNA Replication
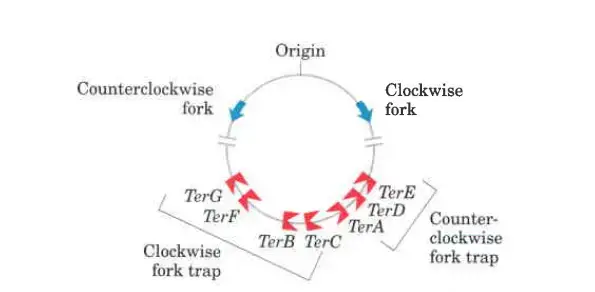
- Ultimately, the circular DNA replication split in two. The two halves of an E. coli chromosome join at a terminus region called Ter, which contains multiple copies of a 20-base-pair sequence.
- The Ter sequences on the chromosome are organised in a way that traps a replication fork inside.
- Protein Tus binds to specific sequences in the Ter RNA (terminus utilisation substance).
- Only one way of a replication fork can be stopped by the Tus-Ter complex. Each replication cycle only allows one Ttrs-Ter complex to work, and that is the complex that is encountered first by one of the two replication forks. Ter sequences may prevent over replication by one fork in the case that the other is delayed or halted by encountering DNA damage or another impediment, even though opposing replication forks typically halt when they intersect.
- So, replication stops when one of the forks runs into a working Tus-Ter complex, and the other fork stops when it runs into the first (arrested) fork.
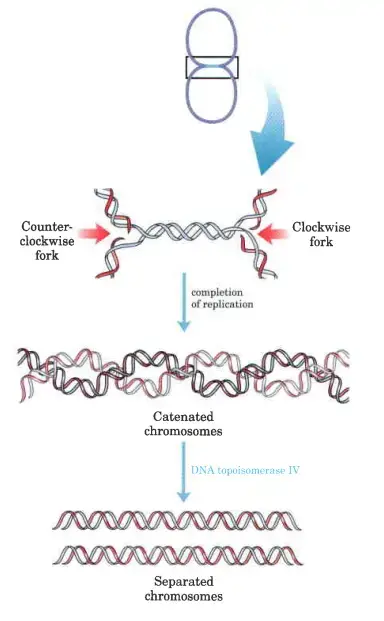
- Following replication of the last few hundred base pairs of DNA (by a mechanism we don’t fully understand), two topologically connected (catenated) circular chromosomes are formed.
- Catenanes are the name given to interconnected DNA cyclins.
- The enzyme topoisomerase IV is necessary for the separation of the catenated rings in E. coLi (a type II topoisomerase).
- Once the chromosomes have been untangled, they are distributed evenly throughout the two daughter cells.
- Many DNA viruses that infect eukaryotic cells undergo a similar process near the end of their replication cycle, which is also characteristic of other circular chromosomes.
Role of topoisomerases in replication termination
During the final stages of DNA replication, the chromosomes are interlinked as catenanes, which are topologically entangled circles of DNA. These catenanes form as a result of the separation of the opposing replication forks. The resolution of these interlinked structures is crucial for ensuring the accurate segregation of chromosomes into daughter cells. Topoisomerases, particularly DNA topoisomerase IV, play a pivotal role in this process. Below is an in-depth explanation of their functions.
1. Formation of Catenanes
- Definition and Structure:
- Catenanes are structures where two or more DNA molecules are interlinked in a manner similar to interlocking rings.
- These structures arise from the process of DNA replication when the newly synthesized strands of DNA from each fork become intertwined.
- Formation Mechanism:
- During replication, the two replication forks move in opposite directions along the DNA molecule.
- As they proceed, they complete the synthesis of new DNA strands, resulting in the formation of interlinked catenanes.
2. Function of Topoisomerases in Resolution
- Topoisomerase IV:
- Type II Topoisomerase:
- DNA topoisomerase IV is a type II topoisomerase found in E. coli and other bacteria.
- It facilitates the resolution of catenanes by transiently breaking both strands of one DNA molecule, allowing the passage of the other linked molecule through the break.
- Mechanism of Action:
- Breaking and Rejoining:
- Topoisomerase IV introduces transient double-strand breaks in one of the catenated chromosomes.
- This temporary cleavage allows the interlinked chromosome to pass through the break.
- After the passage, the enzyme reseals the breaks, thus resolving the catenane structure.
- Breaking and Rejoining:
- Functional Importance:
- Prevents Chromosome Entanglement:
- By resolving catenanes, topoisomerase IV ensures that the chromosomes are separated into individual, unlinked entities.
- This is crucial for proper chromosome segregation during cell division.
- Facilitates Accurate Cell Division:
- The removal of interlinked chromosomes ensures that each daughter cell receives a complete and accurate set of chromosomes.
- Prevents Chromosome Entanglement:
- Type II Topoisomerase:
- Role in Eukaryotic Cells:
- Topoisomerase II:
- In eukaryotic cells, a similar role is performed by topoisomerase II.
- This enzyme also resolves catenanes and other topological constraints that arise during DNA replication and chromosome segregation.
- Topoisomerase II:
3. Implications for Cellular Processes
- Chromosome Segregation:
- Effective resolution of catenanes is essential for accurate chromosome segregation.
- Errors in this process can lead to chromosome missegregation and genomic instability.
- Replication Termination:
- The resolution of catenanes marks the completion of the replication process.
- This step is critical to ensure that replication termination is achieved and that the newly synthesized chromosomes are properly prepared for cell division.
- Drug Targets:
- Antibiotic and Anticancer Therapies:
- Topoisomerases, including topoisomerase IV, are targets for antibiotics and anticancer drugs.
- Inhibitors of these enzymes can effectively disrupt DNA replication and cell division in bacteria and cancer cells.
- Antibiotic and Anticancer Therapies:
Summary
- Topoisomerase IV is crucial in resolving catenanes by transiently breaking and rejoining DNA strands, thereby ensuring proper chromosome segregation and the completion of DNA replication.
- Topoisomerases facilitate the disentangling of interlinked chromosomes, preventing replication errors and maintaining genomic stability.
- Understanding the role of topoisomerases provides insights into cellular processes and highlights potential therapeutic targets for managing bacterial infections and cancer.
Other Prokaryotic replication models
The duplication of theta type has already been mentioned. Rolling-circle replication and D-loop replication are two further forms of bacterial reproduction.
Rolling Circle Replication
- The same circular template DNA spins during bacterial conjugation, and it is around this DNA that a new strand of DNA forms.
- The relaxase enzyme forms a nick in one strand of the conjugative plasmid at the oriT when signalling initiates conjugation.
- Relaxase can function either independently or as part of a larger complex of over a dozen proteins called a relaxosome.
- Relaxase enzyme TraI, along with its cofactors TraY and TraM, and the integrated host factor IHF, make up the relaxosome of the F-plasmid system.
- Following its nicking, the T-strand is unravelled from the intact strand and transmitted to the recipient cell in a 5′-terminus-to-3′-terminus manner.
- In either case, the residual strand undergoes replication, with or without the help of conjugation (conjugative replication similar to the rolling circle replication of lambda phage).
- It’s possible that a second nick is necessary for effective transfer during conjugative replication. The second nicking event is supposedly blocked by compounds that imitate an intermediate step of the process, according to a recent paper.
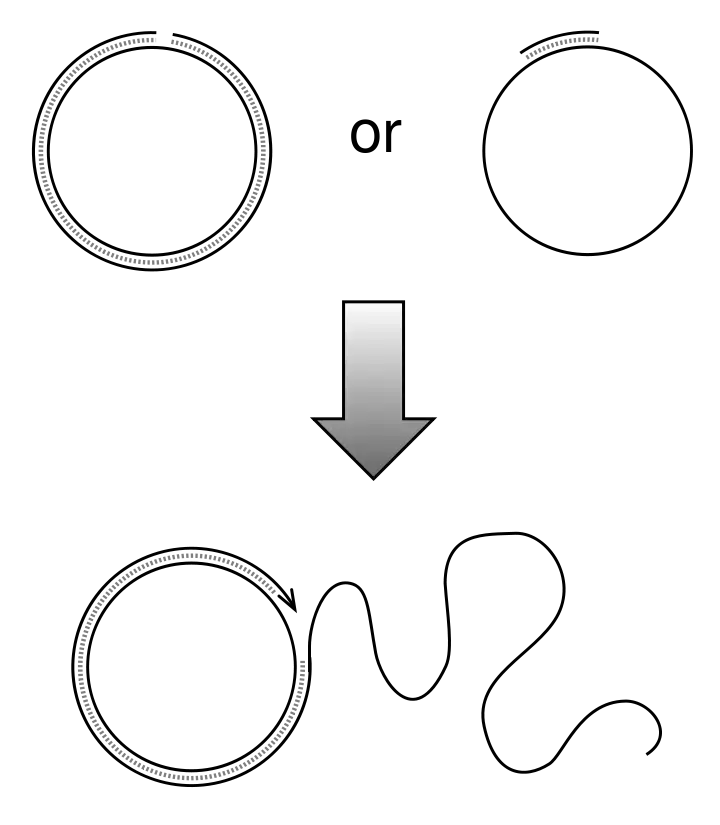
D-loop replication
- Organellar DNA is the most common type of DNA to undergo D-loop replication, which results in the formation of a displacement loop, a triple stranded structure.
Describe The Three-step mechanism of DNA ligation
- DNA ligase I is a cellular enzyme that catalyses the covalent ligation of adenosine monophosphate (AMP) to DNA via hydrolysis of adenosine triphosphate (ATP).
- Following a nick in duplex DNA, the ligase polypeptide transfers an AMP group to the nick’s 5′ phosphate termini.
- In a process involving nucleophilic assault by the 3’HO group and release of AMP, the non-adenylated ligase catalyzes the creation of the phosphodiester link.
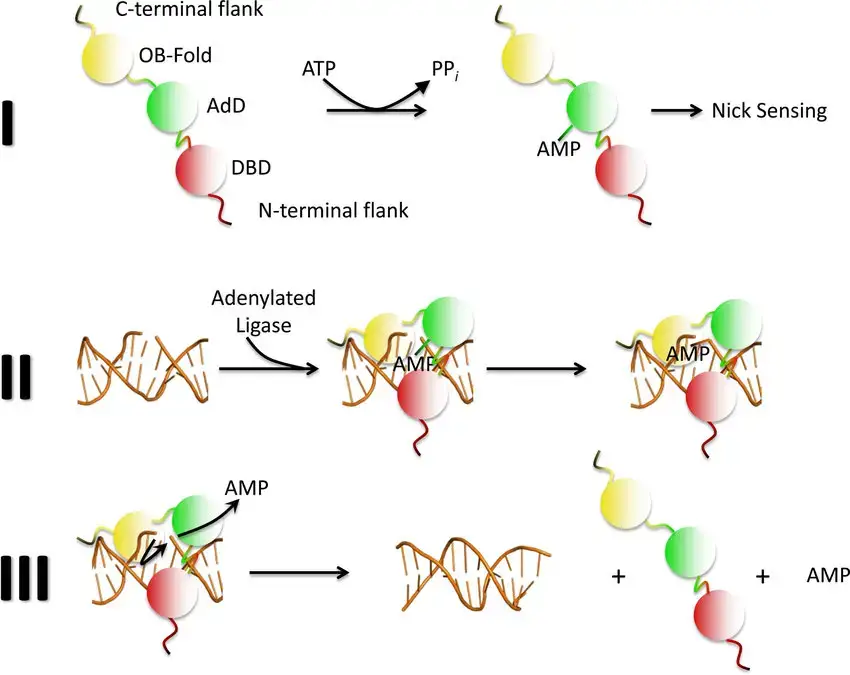
Significance
DNA replication is a critical genetic process essential for cell growth, division, and the conservation of genetic information. This process ensures that each new cell receives an accurate copy of the genome, facilitating proper cellular function and inheritance. Here, we explore the significance of DNA replication in a detailed and sequential manner.
- Foundation of Genetic Continuity
- Genome Conservation:
- DNA replication preserves the entire genome, ensuring genetic information is accurately passed to the next generation.
- This conservation is vital for maintaining the integrity of the species’ genetic blueprint.
- Inheritance:
- Each daughter cell receives an identical copy of the DNA, preserving genetic traits and characteristics through generations.
- Genome Conservation:
- Cell Growth and Division
- Regulation of Cell Cycle:
- DNA replication is tightly regulated to coincide with the cell cycle, ensuring cells only divide when they have a complete set of genetic instructions.
- Proper timing of replication prevents mutations and genomic instability.
- Mitotic and Meiotic Division:
- During mitosis, DNA replication allows for the production of two genetically identical daughter cells, essential for growth and tissue repair.
- In meiosis, replication precedes genetic recombination and reduction division, crucial for sexual reproduction and genetic diversity.
- Regulation of Cell Cycle:
- Molecular Mechanisms and Fidelity
- Enzymatic Precision:
- DNA polymerases and other replication enzymes ensure high-fidelity synthesis of new DNA strands, minimizing errors.
- Proofreading mechanisms correct any mismatches, enhancing the accuracy of replication.
- Replication Fork Dynamics:
- The replication fork, where DNA unwinding and synthesis occur, is orchestrated by a suite of proteins, including helicases, primases, and ligases.
- These proteins ensure the replication process proceeds smoothly and efficiently.
- Enzymatic Precision:
- Impact on Cellular Function
- Protein Synthesis:
- Accurate DNA replication ensures the correct sequence of genes, which is crucial for the synthesis of functional proteins.
- Proper protein synthesis underlies all cellular functions, from metabolism to signaling.
- Cellular Differentiation:
- Replication fidelity allows cells to differentiate correctly, enabling the development of specialized tissues and organs.
- Protein Synthesis:
- Biological Implications
- Development and Growth:
- From a single fertilized egg, DNA replication drives the development of a multicellular organism by enabling successive cell divisions.
- It supports the growth and maintenance of tissues throughout an organism’s life.
- Repair and Regeneration:
- DNA replication plays a role in repairing damaged DNA, maintaining genomic stability, and allowing tissues to regenerate after injury.
- Development and Growth:
FAQ
What is the replication of dna in prokaryotic cells called?
DNA replication in prokaryotes is called theta (theta) replication because this DNA is circular in shape.
What is prokaryotic DNA replication?
Prokaryotic DNA replication is the process by which prokaryotic organisms duplicate their DNA to ensure accurate transmission of genetic information to daughter cells.
What are the three main steps involved in prokaryotic DNA replication?
Prokaryotic DNA replication consists of three steps: initiation, elongation, and termination.
How does initiation of prokaryotic DNA replication occur?
Initiation begins at a specific site called the origin of replication (OriC), where proteins bind to the DNA sequence and helicase unwinds the DNA double helix.
What happens during elongation in prokaryotic DNA replication?
During elongation, DNA polymerase III adds nucleotides one by one to the growing DNA chain, using the parental DNA strands as templates.
What is the role of DNA polymerase III in prokaryotic DNA replication?
DNA polymerase III is the primary enzyme responsible for synthesizing the new DNA strands by adding nucleotides in the 5′-3′ direction.
How is termination of prokaryotic DNA replication achieved?
Termination occurs when the replication forks meet at specific termination sites on the DNA molecule, leading to the separation of the newly synthesized DNA strands.
What is the significance of the origin of replication (OriC) in prokaryotic DNA replication?
OriC is a specific DNA sequence where replication begins. It serves as a recognition site for proteins that initiate the replication process.
What is the function of helicase in prokaryotic DNA replication?
Helicase plays a crucial role in prokaryotic DNA replication by unwinding the DNA double helix, separating the parental strands and creating replication forks.
How are RNA primers involved in prokaryotic DNA replication?
RNA primers, synthesized by the enzyme primase, provide a starting point for DNA synthesis by DNA polymerase. They serve as templates for the addition of DNA nucleotides.
What is the role of DNA ligase in prokaryotic DNA replication?
DNA ligase seals the gaps between Okazaki fragments, which are short DNA segments synthesized on the lagging strand, creating a continuous DNA strand.
References
- Howes, Timothy & Tomkinson, Alan. (2012). DNA ligase I, the replicative DNA ligase. Sub-cellular biochemistry. 62. 327-41. 10.1007/978-94-007-4572-8_17.
- https://www.sciencedirect.com/science/article/pii/S0021925819820197
- https://openoregon.pressbooks.pub/mhccmajorsbio/chapter/dna-replication-in-prokaryotes/
- https://chem.libretexts.org/Courses/Brevard_College/CHE_301_Biochemistry/06%3A_Nucleic_Acids/6.06%3A_DNA_Replication_in_Prokaryotes
- https://pressbooks-dev.oer.hawaii.edu/biology/chapter/dna-replication-in-prokaryotes/
- https://openstax.org/books/biology-2e/pages/14-4-dna-replication-in-prokaryotes
- https://bitesizebio.com/10279/how-dna-ligation-works/
- https://unacademy.com/content/neet-ug/study-material/biology/dna-replication-process-in-prokaryotes/
- https://wou.edu/chemistry/courses/online-chemistry-textbooks/ch450-and-ch451-biochemistry-defining-life-at-the-molecular-level/chapter-9-dna-replication-and-repair-2/
- https://geneticeducation.co.in/prokaryotic-dna-replication-initiation-elongation-and-termination/
- https://www.texasgateway.org/resource/144-dna-replication-prokaryotes
- https://thebiotechnotes.com/2019/07/13/dna-replication-in-prokaryotes/
- https://en.wikipedia.org/wiki/Prokaryotic_DNA_replication
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.