What is Sanger Sequencing?
- Sanger sequencing, a foundational DNA sequencing method, relies on the precise integration of chain-terminating dideoxynucleotides during DNA replication. Developed by Frederick Sanger and his team in 1977, this technique is also known as “dideoxy termination sequencing.”
- Central to Sanger sequencing is the use of dideoxynucleotide triphosphates (ddNTPs). These molecules terminate DNA synthesis because they lack the 3′-OH group necessary for forming a phosphodiester bond with the next nucleotide. DNA polymerase, an enzyme that synthesizes DNA, incorporates these ddNTPs into the growing DNA strand, causing replication to halt. Each of the four ddNTPs (ddATP, ddTTP, ddGTP, ddCTP) is labeled with a different fluorescent dye, enabling the identification of terminated fragments by their final base.
- The process begins with denaturing the double-stranded DNA into single strands, which serve as templates. A primer, complementary to the template’s 3′ end, is then annealed. DNA polymerase extends the primer by adding nucleotides complementary to the template strand. The reaction mixture contains standard deoxynucleotides (dNTPs) and a small proportion of fluorescently labeled ddNTPs. When a ddNTP is incorporated, the extension terminates, creating fragments of varying lengths.
- These fragments are then separated by capillary electrophoresis, a technique that sorts DNA fragments by size. As the fragments pass through a laser, the fluorescent tags are excited, and the emitted light is detected. This detection generates a chromatogram, displaying the sequence of fluorescent signals corresponding to the DNA sequence.
- Sanger sequencing’s accuracy, reaching 99.99%, makes it the gold standard for DNA sequencing. This method played a crucial role in the Human Genome Project, where it was used to sequence small fragments of human DNA, each about 900 base pairs. These fragments were then assembled to reconstruct entire chromosomes, providing a comprehensive map of the human genome.
- Therefore, Sanger sequencing remains a vital tool in molecular biology, genetics, and bioinformatics. Its reliability and precision ensure its continued use for verifying DNA sequences, especially those obtained through next-generation sequencing technologies.
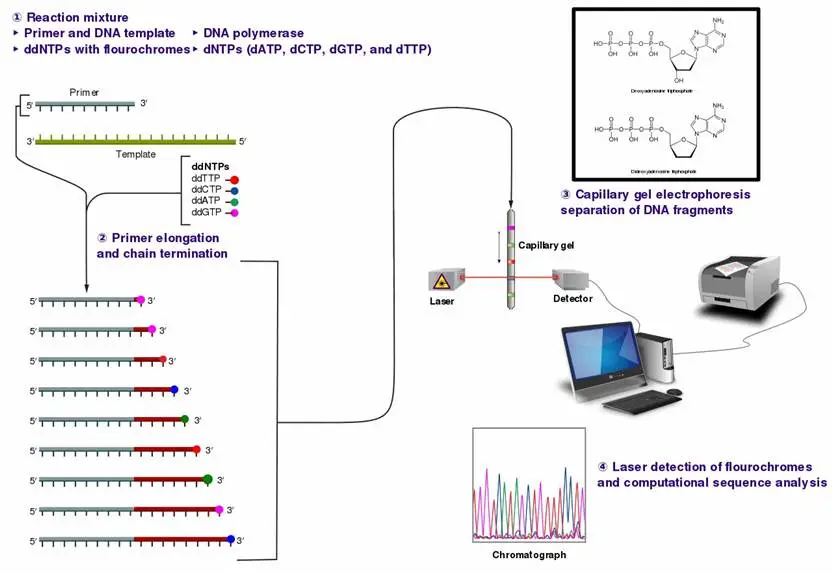
Sanger Sequencing Workflow/Steps
Sanger sequencing is a DNA sequencing method known for its reliability and accuracy. The process involves the formation of a reaction system, amplification of the target segment, gel electrophoresis, and sequence reading. Here’s a step-by-step breakdown of the Sanger sequencing workflow:
1. DNA Sequence for Chain Termination PCR
The workflow begins with the DNA sequence of interest, which serves as a template for chain-termination PCR. This process is similar to standard PCR but includes modified nucleotides called dideoxyribonucleotides (ddNTPs). These ddNTPs are critical because they lack the 3′-OH group necessary for elongation, causing DNA synthesis to terminate when incorporated. Each reaction mixture contains the four standard deoxyribonucleotide triphosphates (dNTPs) and one of the four ddNTPs, each labeled with a distinct fluorescent marker.
- Components of the reaction system:
- DNA template
- DNA polymerase
- Primers
- dNTPs
- ddNTPs (each reaction has a different ddNTP: ddATP, ddTTP, ddCTP, or ddGTP)
2. Size Separation by Gel Electrophoresis
After PCR amplification, the reaction produces a mixture of DNA fragments terminated at various lengths. These fragments are then separated by size using gel electrophoresis.
- Process of gel electrophoresis:
- DNA samples are loaded into a gel matrix.
- An electric current is applied.
- DNA, being negatively charged, moves towards the positive electrode.
- Smaller fragments move faster through the gel, while larger fragments move slower.
This results in a series of bands representing fragments of different lengths.
3. Gel Analysis and Determination of DNA Sequence
The final step involves reading the gel to determine the DNA sequence. Each band corresponds to a DNA fragment terminated by a ddNTP, allowing for the reconstruction of the DNA sequence.
- Reading the gel:
- The shortest fragment terminates at the first nucleotide from the 5′ end.
- The second-shortest fragment terminates at the second nucleotide, and so on.
- By reading the bands from smallest to largest, the sequence of the original DNA strand is determined.
This method provides a clear and accurate readout of the DNA sequence, thanks to the distinctive termination caused by ddNTPs and the precise separation of fragments by gel electrophoresis.
What is Next Generation Sequencing (NGS)?
- Next Generation Sequencing (NGS) is a cutting-edge genetic analysis method that has transformed the field of genomics. It begins by fragmenting genetic material, either DNA or RNA, and attaching known sequence oligonucleotides through adapter ligation. This process allows the fragments to be recognized by the sequencing platform, where the bases in each fragment are identified based on their emission signals.
- Unlike Sanger sequencing, NGS can process millions of sequencing reactions simultaneously. This high-throughput capability offers exceptional sensitivity, rapid results, and cost-effectiveness. Projects that previously took months using traditional methods can now be completed in hours with NGS.
- NGS technologies can be broadly categorized into two approaches: short-read and long-read sequencing. Each approach has distinct advantages and limitations. Short-read sequencing provides high accuracy and coverage but struggles with repetitive regions. Long-read sequencing offers longer read lengths, which helps resolve complex genomic regions, but can have higher error rates.
- The versatility of NGS has driven extensive investment in its development, making it invaluable in both clinical and research settings. This technology, also known as high-throughput sequencing, allows for the accurate, fast, and affordable sequencing of DNA and RNA. Consequently, NGS has become integral to modern molecular biology and genetic research.
- A crucial component of NGS is library preparation. This involves attaching adapters, which include index sequences, barcodes, and sequencing primer binding regions, to the ends of target fragments. These prepared fragments are then sequenced using a dedicated sequencer. Another approach within NGS is targeted sequencing, which selectively captures specific genes or regions of interest before sequencing. Key methods for targeted sequencing include hybrid capture and multiplex PCR.
- Therefore, NGS stands out for its ability to generate vast amounts of genetic data quickly and efficiently. This technological advancement has not only revolutionized genomic research but also expanded our understanding of complex biological systems.
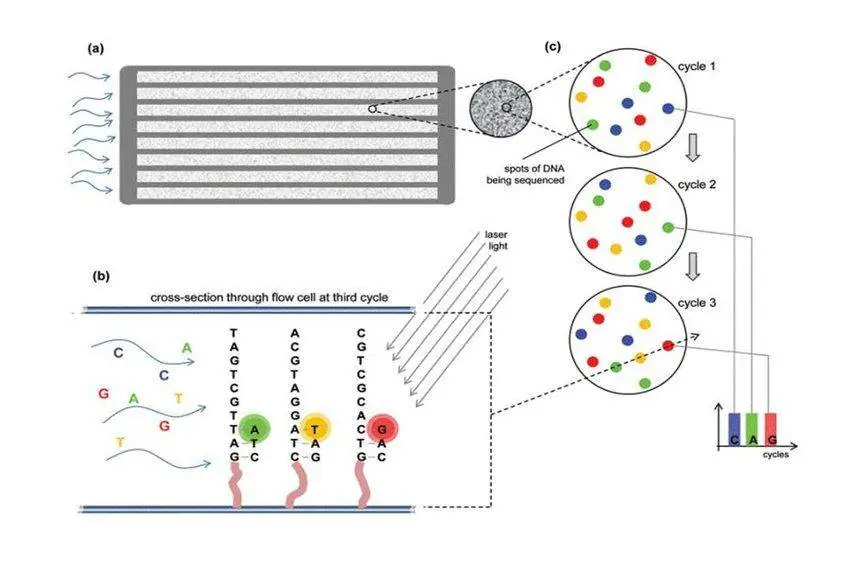
Steps in Next-generation Sequencing Library Preparation
1. Sample Preparation (Pre-treatment)
Nucleic acids, either DNA or RNA, are extracted from selected samples such as blood, sputum, or bone marrow. Quality control (QC) assessment of these extracted samples is crucial and typically involves spectrophotometry, fluorimetry, or gel electrophoresis. If RNA is used, it may need reverse transcription to create cDNA, although some library preparation kits include this step.
2. Fragmentation and Adapter Ligation
The cDNA or DNA is then randomly fragmented using enzymatic treatment or sonication. The optimal fragment length depends on the sequencing platform. Often, a small subset of fragmented samples is run on an electrophoresis gel for optimization. These fragments are end-repaired and ligated to adapters. These adapters are short DNA sequences compatible with the sequencing platform, allowing the fragments to be recognized during multiplex sequencing. Multiplex sequencing uses specific adapter sequences for each sample, enabling the simultaneous sequencing of multiple libraries in one run. The collection of adapter-ligated DNA fragments is called the sequencing library.
3. Size Selection and PCR Amplification
Size selection is performed to remove fragments that are too short or too long for optimal performance on the sequencing platform. This can be done through gel electrophoresis or magnetic bead-based methods. PCR amplification is then used to enrich the library. In emulsion PCR, each fragment attaches to an individual emulsion bead, forming the basis of the sequencing cluster. Post-amplification, a clean-up step using magnetic beads removes unwanted fragments, improving sequencing efficiency.
4. Library Quality Control
The final library undergoes quality control using quantitative PCR (qPCR) to verify the quality and quantity of DNA. This ensures that samples are prepared at the correct concentration for sequencing.
5. Sequencing
Depending on the platform, clonal amplification of library fragments occurs either before loading the sequencer (PCR) or on the sequencer itself (bridge PCR). The sequencer then detects and records the sequences.
6. Data Analysis
The generated data files are analyzed according to the specific workflow and study objectives. Analysis methods vary greatly depending on the goals of the study.
Paired-end and paired-pair sequencing, while reducing the number of samples analyzed in a single run, offer advantages in downstream data analysis. These methods involve sequencing reads from both ends of a fragment (paired-end) or those separated by interstitial DNA regions (paired pairs), aiding in de novo assembly and other complex analyses.
Sanger Sequencing vs NGS (Sanger Sequencing vs Next Generation Sequencing)
Sanger Sequencing and Next-Generation Sequencing (NGS) represent two cornerstone technologies in genomic analysis. Each has distinct principles, applications, and advantages. Here, we compare the two methods in a detailed and sequential manner.
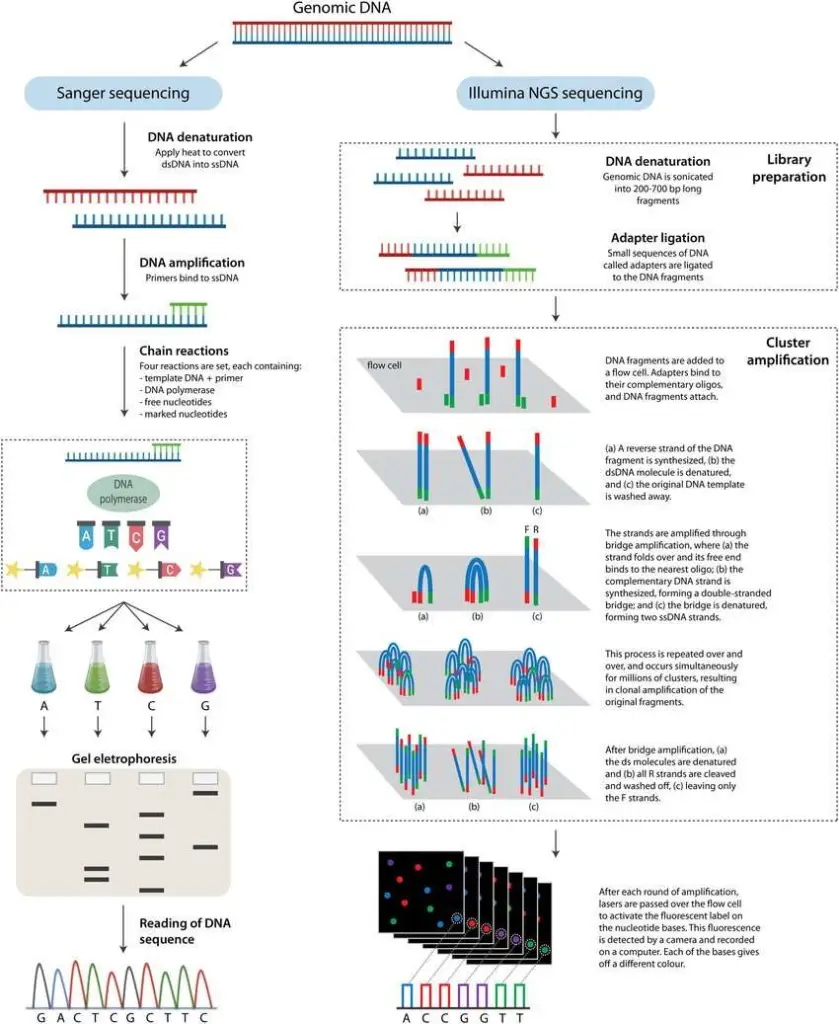
Sequencing Principle
- Sanger Sequencing: This method relies on the chain termination technique. It incorporates chain-terminating nucleotides (ddNTPs) to halt DNA synthesis at specific points, generating DNA strands of varying lengths.
- Next-Generation Sequencing (NGS): NGS uses parallel sequencing technologies to simultaneously sequence numerous DNA fragments, producing vast amounts of sequence data in a single run.
Read Length
- Sanger Sequencing: Typically produces longer reads, often reaching 800-1000 base pairs.
- NGS: Read lengths vary by platform and application but tend to be shorter. For instance, the Illumina platform produces short sequences.
Speed
- Sanger Sequencing: Relatively slow, as it sequences small DNA segments one at a time, requiring sequential analysis.
- NGS: Significantly faster due to its high-throughput nature, processing multiple samples concurrently.
Cost
- Sanger Sequencing: Relatively high, especially for large-scale projects.
- NGS: More cost-effective for high-throughput projects, reducing the per-sample cost significantly.
Error Rate
- Sanger Sequencing: Low error rate, known for its accuracy.
- NGS: Higher error rates, but errors can be mitigated through repeated sequencing and advanced statistical methods.
Applications
- Sanger Sequencing: Ideal for small-scale projects, such as SNP identification and gene cloning.
- NGS: Suitable for large-scale applications, including whole-genome sequencing, transcriptome sequencing, and protein-nucleic acid interaction studies.
Equipment Cost
- Sanger Sequencing: Lower equipment costs due to simpler instrumentation.
- NGS: Higher equipment costs, requiring complex and sophisticated devices.
Scope of Application
- Sanger Sequencing: Best suited for small DNA fragments.
- NGS: Effective for large-scale projects, including whole-genome and whole-transcriptome sequencing.
Parallelization
- Sanger Sequencing: Sequential, allowing only one DNA strand to be sequenced at a time.
- NGS: Highly parallel, processing multiple samples or fragments simultaneously, enhancing efficiency.
Sample Handling
- Sanger Sequencing: Efficient for individual samples, less suited for high-throughput processing.
- NGS: Designed for high-throughput sequencing, handling multiple samples simultaneously.
Specificity
- Sanger Sequencing: Exhibits good specificity with minimal cross-contamination.
- NGS: Potential for cross-contamination in high-throughput settings, requiring additional processing steps.
Data Analysis
- Sanger Sequencing: Straightforward analysis, ideal for small-scale projects.
- NGS: Requires sophisticated tools and algorithms to manage and interpret large-scale data.
Integration of New Technologies
- Sanger Sequencing: A mature technology with limited adaptability to new advancements.
- NGS: Highly adaptable, continuously incorporating new sequencing technologies and methodologies.
Availability
- Sanger Sequencing: Widely used in academic research for many years.
- NGS: Extensively applied across academic, medical, and industrial research settings.
| Feature | Sanger Sequencing | Next-Generation Sequencing (NGS) |
|---|---|---|
| Sequencing Principle | Chain termination method, using ddNTPs to halt DNA synthesis. | Parallel sequencing of multiple DNA fragments, producing extensive data. |
| Read Length | Long reads, typically 800-1000 base pairs. | Shorter reads, varying by platform (e.g., Illumina produces short reads). |
| Speed | Relatively slow, sequencing segments sequentially. | Fast, high-throughput, processing multiple samples concurrently. |
| Cost | Relatively high for large-scale projects. | More economical for high-throughput projects. |
| Error Rate | Low, highly accurate. | Higher error rate, but correctable with repeated sequencing. |
| Applications | Small-scale projects, SNP identification, gene cloning. | Large-scale genome sequencing, transcriptome sequencing, etc. |
| Equipment Cost | Lower, simpler equipment. | Higher, requiring sophisticated instruments. |
| Scope of Application | Small DNA fragments. | Large-scale, whole-genome, or whole-transcriptome sequencing. |
| Parallelization | Sequential, one DNA strand at a time. | Parallel, multiple samples or fragments at once. |
| Sample Handling | Effective for individual samples, less suited for high-throughput. | Suitable for high-throughput, processing multiple samples simultaneously. |
| Specificity | Good specificity, minimal cross-contamination. | Potential for cross-contamination, requires additional steps. |
| Data Analysis | Straightforward, suitable for small-scale projects. | Requires advanced tools and algorithms for large-scale data. |
| Integration of New Technologies | Mature, less adaptable. | Highly adaptable, incorporating new technologies. |
| Availability | Widely used in academic research. | Extensively applied in academic, medical, and industrial research. |
Benefits and Limitations of Sanger Sequencing vs. Next-Generation Sequencing (NGS)
Benefits
Sanger Sequencing:
- High Accuracy and Reliability: Sanger sequencing is renowned for its accuracy, producing highly reliable data. This method is less prone to errors compared to some high-throughput techniques.
- Established Workflow: The protocols for Sanger sequencing are well-established and widely used. Researchers are familiar with the process, making it easier to implement in many laboratories.
- Higher Sequencing Depth: Sanger sequencing provides higher sequencing depth, which is beneficial for detecting variants with high precision.
Next-Generation Sequencing (NGS):
- High Throughput and Scalability: NGS can sequence millions of fragments simultaneously, making it highly efficient for large-scale projects. This parallel processing capability significantly reduces sequencing time.
- Superior Sensitivity: NGS is highly sensitive and can detect low-frequency variants that might be missed by Sanger sequencing. This sensitivity is crucial for identifying rare mutations.
- Comprehensive Genomic Coverage: NGS offers extensive coverage of the genome, allowing for the analysis of large, complex regions. It is ideal for whole-genome and whole-transcriptome sequencing.
Limitations
Sanger Sequencing:
- Lower Throughput: Sanger sequencing processes one fragment at a time, making it less suitable for large-scale projects. This sequential nature limits the speed and volume of data production.
- Higher Costs for High-Volume Targets: The cost of Sanger sequencing can be prohibitive for projects requiring the sequencing of numerous or large DNA fragments.
- Limited Discovery Power: Compared to NGS, Sanger sequencing has limited capacity for discovering new variants, especially in large and complex genomic regions.
Next-Generation Sequencing (NGS):
- Initial Setup and Data Analysis Complexities: NGS requires sophisticated equipment and complex data analysis tools. The initial setup can be challenging, and analyzing the large volumes of data generated necessitates advanced computational resources.
- Cost Considerations for Small-Scale Projects: While NGS is cost-effective for large-scale sequencing, it can be expensive for small-scale projects. The high cost of reagents and instruments may not be justifiable for smaller studies.
- Potential for Sequencing Errors: NGS has a higher error rate compared to Sanger sequencing. Although these errors can be mitigated through repeated sequencing and advanced statistical methods, they still pose a challenge for data accuracy.
| Aspect | Sanger Sequencing | Next-Generation Sequencing (NGS) |
|---|---|---|
| Benefits | – Demonstrates high accuracy and reliability | – Exhibits high throughput and scalability |
| – Embraces a familiar workflow with established protocols | – Offers superior sensitivity for low-frequency variants | |
| – Provides higher sequencing depth for variant detection | – Encompasses comprehensive genomic coverage | |
| Limitations | – Exhibits lower throughput, sequencing one fragment at a time | – Presents initial setup and data analysis complexities |
| – Incurs higher costs for high-volume targets | – Entails cost considerations for small-scale projects | |
| – Demonstrates limited discovery power compared to NGS | – Carries a potential for sequencing errors and artifacts |