Polyploidy plays a pivotal role in plant breeding, especially in enhancing the genetic diversity and adaptability of crops. Polyploidy refers to the condition in which a plant has more than two complete sets of chromosomes, a state that naturally occurs due to irregularities in mitotic or meiotic divisions. These irregularities can lead to an increase in the chromosome number beyond the diploid state, resulting in polyploid individuals. Polyploidy is particularly valuable because it can create novel genetic combinations and traits that contribute to crop improvement.
There are two primary types of polyploidy: autopolyploidy and allopolyploidy. Autopolyploids arise when chromosome duplication occurs within the same species, leading to multiple copies of the same genome. Allopolyploids, on the other hand, occur when chromosomes from different species are combined, usually through hybridization followed by chromosome doubling. This latter type of polyploidy is especially important in plant breeding as it facilitates the merging of desirable traits from two different species, thereby overcoming the sterility that often results from hybridization.
Polyploid plants tend to exhibit characteristics that are advantageous in agriculture. For instance, they often have increased cell size, which can lead to larger fruits, seeds, and leaves. Furthermore, polyploidy can confer greater resilience to environmental stresses such as drought and disease. This is due to the redundancy in genetic material, which provides plants with greater flexibility to adapt to challenging conditions.
Inducing polyploidy in plants can be achieved through chemical treatments, with colchicine being a commonly used agent. Colchicine disrupts the normal spindle fiber formation during cell division, preventing the separation of chromosomes and thus doubling the chromosome number in the resulting cells. Polyploidy can also occur spontaneously in nature, contributing to the natural evolution of plants. Many of the world’s most important crops, such as wheat, cotton, and potatoes, are polyploids, either naturally or as a result of human intervention.
In plant breeding, polyploidy offers unique opportunities to overcome the limitations of diploid genetics. By creating polyploid plants, breeders can introduce genetic variability, improve stress tolerance, and increase productivity. Besides, polyploidy allows for the combination of traits from distant species, which can be challenging to achieve through traditional breeding methods alone. Therefore, polyploidy not only enhances crop characteristics but also expands the scope of genetic improvement in agriculture.
What Is Polyploidy?
- Polyploidy refers to a condition in which an organism’s cells possess more than two complete sets of chromosomes. This contrasts with the diploid state, where cells contain two complete sets, one from each parent, making a total of two homologous chromosomes per set. Polyploidy is particularly prevalent in plants and can occur through various mechanisms during the evolution of species.
- Polyploidy arises from whole-genome duplication, which can result from abnormal cell division. This process may occur during mitosis or more commonly during meiosis when chromosomes fail to separate properly. Additionally, polyploidy can be induced artificially in plants and cell cultures using chemicals such as colchicine and oryzalin, which facilitate chromosome doubling.
- Among animals, polyploid cells are found in specific organs such as the brain, liver, heart, and bone marrow. Polyploidy is also observed in species like goldfish, salmon, and salamanders. In plants, polyploidy is widespread and includes both wild and cultivated species. For instance, wheat exhibits varying ploidy levels, including diploid (two sets), tetraploid (four sets), and hexaploid (six sets). Similarly, many species within the Brassica genus are tetraploids, and sugarcane can exceed octaploidy.
- Polyploidization can lead to sympatric speciation, where new species arise in the same geographic area as their ancestors and are typically unable to interbreed with them. An example of this is Erythranthe peregrina, which originated from a triploid hybrid and developed through genome duplication.
- Conversely, polyploidy can also facilitate ‘reverse speciation,’ where gene flow between previously isolated lineages is enabled. This phenomenon has been observed in Arabidopsis arenosa and Arabidopsis lyrata, where polyploidy events allowed adaptive gene flow between species.
- Polyploid organisms are characterized by having more than two sets of genomes. In many plant genera, such as Rosa (roses), species exhibit a range of ploidy levels including diploid, triploid, tetraploid, pentaploid, hexaploid, and octaploid configurations. Although natural populations rarely exceed tetraploidy, polyploidy is common in agriculture and ornamental plants, such as hexaploid wheat and octaploid strawberries.
- In general, polyploid plants often display larger organ sizes and enhanced resistance to environmental stress compared to their diploid counterparts. Polyploidy thus plays a significant role in plant evolution and crop improvement, contributing to the diversity and adaptability of plant species.
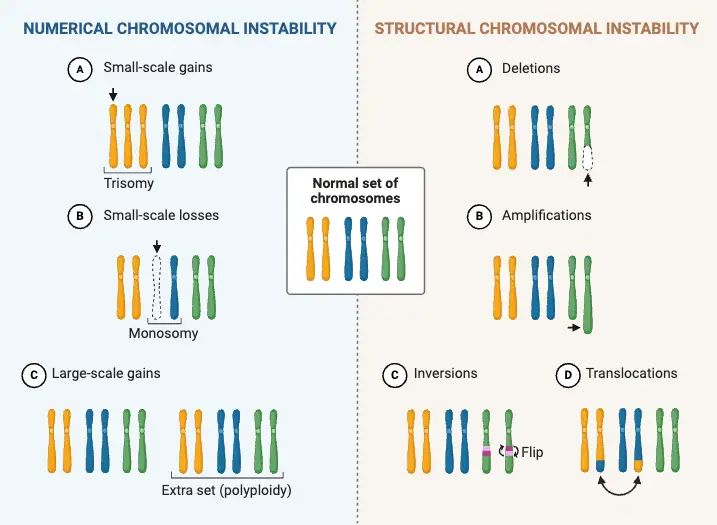
Types of Changes in Chromosome Number
Changes in chromosome number can profoundly influence the genetic and phenotypic characteristics of plants. These alterations are categorized into two primary groups: aneuploidy and euploidy. Each type of chromosomal change has specific characteristics, implications, and applications in plant breeding and research.
1. Aneuploidy
Definition: Aneuploidy is characterized by a chromosome number that deviates from the exact multiple of the basic chromosome set (x). This condition involves gains or losses of individual chromosomes but does not involve the entire set.
Types of Aneuploidy:
- Nullisomics (2n – 2):
- Description: Nullisomics lack an entire chromosome pair.
- Applications: Useful for studying the effects of missing chromosomes on plant development and understanding gene function.
- Monosomics (2n – 1):
- Description: Monosomics have one chromosome missing from a pair.
- Applications: Employed in breeding programs to investigate gene linkage and create detailed chromosome maps.
- Double Monosomics (2n – 1 – 1):
- Description: These individuals are missing two chromosomes, each from different pairs.
- Applications: Used to study the combined effects of missing chromosomes on plant traits.
- Trisomics (2n + 1):
- Description: Trisomics possess an extra chromosome.
- Applications: Investigate gene dosage effects and are sometimes utilized to produce specific phenotypic variations.
- Double Trisomics (2n + 1 + 1):
- Description: These individuals have two additional chromosomes, each from different chromosome pairs.
- Applications: Useful for studying the combined effects of additional chromosomes on plant development.
- Tetrasomics (2n + 2):
- Description: Tetrasomics contain an extra pair of chromosomes.
- Applications: Valuable in research on chromosome behavior and inheritance patterns.
2. Euploidy
Definition: Euploidy refers to changes where the chromosome number is an exact multiple of the basic chromosome number (x). It is commonly associated with polyploidy, where there are additional complete sets of chromosomes.
Types of Euploidy:
- Monoploids (x):
- Description: Monoploids have a single set of chromosomes.
- Applications: Rare in nature, but useful for mutation breeding and genetic analysis.
- Diploids (2x):
- Description: The standard chromosome number for many species, with two sets of chromosomes.
- Applications: Represents the normal state for most organisms and is the baseline for comparing other ploidy levels.
- Polyploids:
- Description: Individuals with more than two sets of chromosomes. Polyploidy is significant in plant evolution and breeding.
- Types:
- Triploids (3x):
- Description: Contain three sets of chromosomes.
- Applications: Often sterile but may exhibit desirable traits such as increased fruit size.
- Tetraploids (4x):
- Description: Have four sets of chromosomes.
- Applications: More stable and fertile than triploids. Commonly used in breeding programs to enhance crop traits.
- Pentaploids (5x), Hexaploids (6x), and Beyond:
- Description: Individuals with five or more sets of chromosomes.
- Applications: Higher-level polyploids, such as hexaploids in wheat, contribute to the genetic diversity and adaptability of crops.
- Triploids (3x):
- Allopolyploids:
- Description: Formed through hybridization between different species, combining distinct genomes.
- Applications: Create species with new traits by combining genetic material from different sources. For example, allotetraploids have two distinct sets of genomes, leading to novel and useful traits.
- Amphidiploids:
- Description: A type of allopolyploid where the individual behaves as a diploid during meiosis due to having two copies of each genome.
- Applications: Crucial for producing fertile hybrids between species. They help bridge genetic gaps and facilitate breeding programs.
Historical Milestones in Heteroploidy Research
The history of heteroploidy, which refers to variations in chromosome number, began in the early 20th century and has played a significant role in understanding plant genetics and evolution. Various experiments led to the discovery of changes in chromosome numbers, contributing to the development of new species and advancements in plant breeding.
Key Historical Milestones in Heteroploidy Research:
- First Observation of Heteroploidy (1907)
- The first recorded instance of heteroploidy was the discovery of the “gigas” mutant in Oenothera by Lutz in 1907. This mutant was an autotetraploid (4n), marking the first observation of polyploidy in a natural population.
- Discovery of Trisomy (1910)
- In 1910, Blakeslee discovered the “globe” mutant in Datura stramonium (dhatura), which was later confirmed by Belling in 1920 to be trisomic. This was the first documented case of aneuploidy, a condition where an individual has an abnormal number of chromosomes, either missing or having extra chromosomes.
- First Experimental Induction of Autotetraploidy (1916)
- Winkler, in 1916, was the first to experimentally induce autotetraploidy in Solanum nigrum (black nightshade). By decapitating shoots, some of the regenerating shoot buds became tetraploid, marking a breakthrough in manipulating chromosome numbers for experimental purposes.
- Interspecific Hybridization and Chromosome Doubling Hypothesis (1917)
- Winge, in 1917, proposed that interspecific hybridization followed by chromosome doubling could play a significant role in the evolution of new species. This hypothesis laid the groundwork for future synthetic species development and hybridization research.
- First Synthetic Species: Triticale (1890, Studied in 1936)
- The first experimentally synthesized species was Triticale, produced by Rimpau in 1890 through a cross between Triticum (wheat) and Secale (rye). However, its chromosome number was not studied until 1936, delaying full understanding of its genetic structure.
- First Fully Studied Synthetic Species (1925)
- The first synthetic species with complete chromosomal information was Nicotiana digluta, which resulted from a spontaneous chromosome doubling of the F1 hybrid between Nicotiana glutinosa and Nicotiana tabacum. This species was synthesized by Clausen and Goodspeed in 1925, highlighting the potential for synthetic speciation through chromosome manipulation.
- Discovery of Colchicine’s Chromosome Doubling Effect (1937)
- Colchicine, a chemical that induces chromosome doubling, was independently discovered by Blakeslee and Nebel in 1937. This discovery revolutionized the ability to artificially induce polyploidy in plants. Colchicine disrupts mitosis, leading to the failure of chromosome separation, thereby doubling the chromosome number in treated cells.
- Further Insights into Colchicine and Mitosis (1955)
- Although colchicine’s chromosome doubling effect was understood in 1937, its precise impact on mitosis was not fully explored until 1955. This later research provided a deeper understanding of how the drug interferes with spindle fiber formation, allowing scientists to better control polyploidy induction.
Aneuploidy
Aneuploidy refers to a deviation from the normal chromosome number in a cell, where the chromosome set is incomplete or contains additional chromosomes. Unlike euploidy, where chromosome numbers change in exact multiples of the basic set, aneuploidy involves the gain or loss of one or more individual chromosomes. This condition plays a significant role in genetic studies, particularly in understanding plant breeding and evolution.
Types of Aneuploidy
- Monosomy (2n – 1)
- Monosomics are individuals lacking a single chromosome. They are particularly useful in genetic studies for mapping traits in polyploid species, such as wheat. However, monosomy may not always be viable in some species, especially if the missing chromosome carries essential genes.
- Nullisomy (2n – 2)
- Nullisomics are individuals missing an entire chromosome pair. This form of aneuploidy is rare and typically only viable in highly polyploid species like wheat and oats. In species with lower ploidy levels, such as tobacco, nullisomics are generally not viable due to the absence of critical genetic material.
- Trisomy (2n + 1)
- Trisomics have an additional chromosome and are classified into three types:
- Primary trisomic: The extra chromosome is identical to one of the chromosomes from the haploid genome.
- Secondary trisomic: The extra chromosome is an isochromosome, meaning both arms of the chromosome are identical.
- Tertiary trisomic: The additional chromosome is translocated from another chromosome set.
- Trisomics have an additional chromosome and are classified into three types:
- Tetrasomy (2n + 2)
- Tetrasomics possess an extra pair of homologous chromosomes. These plants produce a considerable frequency of n+1 gametes, which increases the likelihood of trisomic offspring in their progeny.
Mechanisms of Aneuploidy Production
- Spontaneous Aneuploidy
- Aneuploid individuals can arise spontaneously due to meiotic irregularities. For example, in Datura, about 0.4% of the pollen is aneuploid (n+1), leading to spontaneous formation of aneuploid gametes. These gametes may fuse with normal gametes, producing aneuploid progeny.
- Asynaptic and Desynaptic Plants
- In asynaptic and desynaptic plants, chromosomes fail to pair properly during meiosis, leading to the formation of univalents (unpaired chromosomes). This irregular pairing often results in a high frequency of aneuploid progeny, as chromosomes may segregate unevenly.
- Translocation Heterozygotes
- In plants that are heterozygous for a translocation, chromosomes can form a ring or chain of four during meiosis. A 3:1 disjunction of these structures may result in one n+1 gamete and one n-1 gamete, leading to the production of aneuploid offspring with variable chromosome numbers.
- Tetrasomic Plants
- Tetrasomics (2n+2) produce a high frequency of n+1 gametes. These plants are valuable for generating trisomic individuals, which are useful for genetic studies. In controlled breeding programs, tetrasomics are often maintained for the systematic production of trisomics.
Morphological and Cytological Features of Aneuploids
Aneuploidy, which involves the addition or loss of chromosomes from the normal set, significantly affects both the morphology and cytology of plants. These deviations from the typical diploid condition introduce various changes that impact the organism’s vigor, fertility, and overall development.
Morphological Features:
- Weaker Growth
- Aneuploid plants are generally weaker compared to their diploid counterparts. This reduced vigor is evident in several species, particularly in monosomics and nullisomics. For example, monosomics, where one chromosome is missing, often do not survive in diploid species. Nullisomics, which lack an entire chromosome pair, rarely survive in polyploid species like tobacco. However, exceptions exist, such as the wheat variety Chinese Spring, which can sustain all 21 nullisomics.
- Variation in Trisomic Plants
- Trisomics, where an extra chromosome is present, show varied levels of vigor depending on the chromosome involved. In some cases, such as trisomic 5 in maize, the plants may resemble normal diploids in vigor. However, many trisomics exhibit significantly reduced strength and abnormal growth. The type of chromosomal gain or loss often determines the extent of morphological alteration.
- Distinct Morphological Effects
- Aneuploid plants tend to display distinct morphological traits due to the irregular chromosome number. Almost all features of the plant are affected, although morphology is not always a reliable indicator of the specific chromosomal change. For instance, while monosomics in wheat can be morphologically similar to diploids, the underlying chromosomal imbalance may not be evident without cytological analysis.
Cytological Features:
- Behavior in Meiosis
- Cytologically, aneuploids exhibit irregular chromosome behavior during meiosis, particularly during metaphase I and anaphase I.
- Monosomics generally show one unpaired chromosome, known as a univalent, during metaphase I. This univalent can exhibit various behaviors, such as moving to one of the poles, lagging, being lost, or dividing into chromatids like in mitosis. The irregular movement of the univalent causes monosomics to produce a higher proportion of gametes with one less chromosome (n-1).
- Nullisomics, in contrast, typically show regular bivalent formation and gamete production, despite the absence of a chromosome pair. Their meiotic behavior allows for relatively normal gamete production, albeit with n-1 chromosome numbers.
- Tetrasomics, with an extra pair of chromosomes, often form quadrivalents, where four homologous chromosomes pair together during metaphase I. These quadrivalents usually separate evenly (2:2) at anaphase I, but irregular separations can occur, leading to the production of both n and n+1 gametes.
- Trisomics form trivalents, where three homologous chromosomes pair together. The behavior of trivalents during meiosis is less predictable than bivalents, resulting in the production of a smaller proportion of n+1 gametes, typically less than 50%.
- Cytologically, aneuploids exhibit irregular chromosome behavior during meiosis, particularly during metaphase I and anaphase I.
- Transmission and Fertility
- The transmission of aneuploidy is often inefficient, particularly through pollen grains. The slower growth of aneuploid pollen tubes compared to their diploid counterparts reduces the success rate of fertilization. Moreover, aneuploid seeds tend to be smaller, show lower germination rates, and exhibit reduced seedling viability. These factors contribute to the overall reduction in the recovery of aneuploid progeny.
- Seed and Gamete Viability
- Due to irregular chromosome segregation during meiosis, aneuploids often produce a disproportionate number of abnormal gametes (n-1 or n+1). This leads to reduced seed viability, smaller seed size, and lower rates of successful germination. The survival rate of aneuploid offspring is also compromised due to these cytological abnormalities.
Applications of Aneuploids in Crop Improvement
The following points outline the key applications of aneuploids in crop improvement:
- Understanding Gene Dosage Effects
- Function: Aneuploids are instrumental in studying the effects of losing or gaining entire chromosomes or chromosome arms on the phenotype of an individual. By examining how these chromosomal alterations affect plant traits, researchers can better understand the balance of genes required for normal development.
- Application: This understanding aids in identifying how specific genes or chromosomal regions contribute to phenotypic traits, allowing breeders to develop crops with desirable characteristics by manipulating gene dosage.
- Locating Genes on Chromosomes
- Function: Aneuploids, especially secondary and tertiary trisomics, are used to map genes to specific chromosomes or even to particular chromosome arms. This technique helps in pinpointing the exact location of genes within a chromosome.
- Application: This precise gene mapping is crucial for gene discovery and for developing molecular markers that can be used in marker-assisted selection to improve crop varieties.
- Studying Chromosomal Homoeology
- Function: The study of aneuploids has revealed insights into chromosomal homoeology, particularly in wheat. For instance, the presence of a tetrasomic condition in one genome can compensate for the absence of the corresponding chromosome in another genome, maintaining normal plant development.
- Application: This knowledge helps in understanding the genetic relationships between different genomes and assists in breeding programs aimed at improving genetic stability and yield in crops like wheat.
- Identifying Chromosomes Involved in Translocation
- Function: Aneuploids are valuable for identifying chromosomes involved in genetic translocations, where segments of chromosomes are swapped between non-homologous chromosomes.
- Application: This identification is crucial for understanding the genetic basis of traits affected by translocations and for creating crops with novel genetic combinations that may confer beneficial traits such as disease resistance or stress tolerance.
- Producing Substitution Lines
- Function: Aneuploids facilitate the creation of substitution lines, where one or more chromosomes from one variety are substituted into another variety. This technique allows researchers to study the effects of individual chromosomes or to transfer specific genes between varieties.
- Application: Substitution lines are useful for exploring the functional roles of different chromosomes in a variety and for transferring beneficial traits from one variety to another, enhancing crop performance and adaptability.
Aneuploid Analysis for Locating Genes on Particular Chromosomes
Aneuploid analysis, an essential tool in genetics, is instrumental in locating specific genes on particular chromosomes. By utilizing various forms of aneuploidy, such as nullisomic, monosomic, or trisomic analysis, researchers can map genes and understand their chromosomal positions. This method is particularly valuable in plant breeding and crop improvement, enabling scientists to target and manipulate desired genetic traits.
1. Nullisomic Analysis
Nullisomic analysis is used in polyploid species like wheat, where the absence of a chromosome allows researchers to identify the chromosomal location of specific genes. There are four key approaches used in nullisomic analysis:
- Absence of Expression of a Dominant Character:
If a dominant gene is located on a specific chromosome, its trait will not be expressed in a nullisomic individual lacking that chromosome. For example, genes responsible for seed color or awn inhibition in wheat have been mapped using this method. - Study of the F₁ Generation:
In this approach, a strain carrying the dominant gene is crossed with a complete set of nullisomics. The resulting F₁ progeny, primarily monosomic, are further analyzed. The chromosome contributing the unpaired gene is identified based on the segregation of the dominant and recessive traits in the next generation (F₂). - Study of F₃ Progenies:
F₃ generation studies help confirm gene location through further analysis of disomic plants. In families where the critical gene is located, a uniform expression of the dominant trait will occur, while other families will show a 3:1 segregation ratio. - Chromosome Substitution:
Nullisomic analysis facilitates chromosome substitution, where individual chromosomes are transferred between varieties to study their genetic effects in a common background or to transfer desirable genes. This is particularly useful in creating varieties with improved traits by swapping chromosomes carrying beneficial genes.
2. Monosomic Analysis
Monosomic analysis, applied primarily in diploid and polyploid species, is a valuable method for gene localization. This technique was first used in Nicotiana tabacum and later extended to other crops. It involves the following key processes:
- Crossing with a Dominant Gene:
A monosomic line, identified through cytological analysis, is crossed with a strain carrying a dominant gene. The resulting F₁ progeny are analyzed, and the plants that exhibit the dominant trait indicate the chromosome responsible for carrying the gene. - Selfing of Monosomic Plants:
The F₁ monosomic plants are selfed to produce an F₂ generation, where plants lacking the dominant trait are identified as nullisomic. In crops like tobacco, where nullisomics do not survive, the absence of such plants in the progeny confirms the gene’s chromosomal location.
3. Trisomic Analysis
Trisomic analysis is applied in diploid species where nullisomic and monosomic analysis is not feasible. It involves crossing a strain carrying a gene of interest with trisomic plants, and then analyzing the F₁ and subsequent generations. The following key steps are involved:
- Crossing with Trisomic Plants:
In this process, the F₁ generation includes both trisomic and disomic plants, with the disomics being discarded. Trisomic F₁ plants are either selfed or test-crossed to a recessive disomic strain. - Segregation Analysis:
The F₂ or test-cross progeny are evaluated for their trait expression. A typical 3:1 or 1:1 segregation ratio would be expected if the gene is not located on the trisomic chromosome. However, significant deviations from these ratios indicate that the gene resides on the extra chromosome in the trisomic state. This method has been successfully applied in crops like maize, barley, and tomato.
Limitations of Aneuploid Analyses
Aneuploid analyses, despite their significant contributions to genetic research and crop improvement, face several inherent limitations:
- Production and Maintenance Complexity
- Requirement for a Complete Set: Creating and sustaining a comprehensive set of aneuploids is essential for accurate analysis. This process involves considerable effort and technical skill.
- Challenges in Production: The production of aneuploids demands precise cytogenetic techniques, which are both time-consuming and technically demanding.
- Maintenance Issues: Maintaining these aneuploids is further complicated by the phenomenon of univalent shift. This shift occurs when progeny of an aneuploid plant exhibit aneuploidy for chromosomes other than those of the original aneuploid parent. This variability necessitates rigorous monitoring and management.
- Univalent Shift
- Definition and Impact: Univalent shift refers to the alteration in chromosome number in progeny from the original aneuploid plant. Specifically, it involves the formation of univalents in chromosomes other than the ones originally involved in aneuploidy.
- Cytogenetic Implications: This phenomenon can obscure results and complicate the interpretation of genetic data, requiring additional cytological analysis to ensure accuracy.
- Requirement for Extensive Cytological Analysis
- Time-Consuming Procedures: Accurate aneuploid analysis necessitates detailed cytological work. This includes frequent chromosome counting and the identification of aneuploid states, which is labor-intensive.
- Technical Skill and Accuracy: High proficiency in cytogenetics is required to conduct and interpret these analyses correctly. Any errors or inaccuracies in cytological work can lead to erroneous conclusions about the genetic makeup and phenotypic expressions of the aneuploids.
- Challenges in Chromosome Substitution
- Complexity in Execution: Chromosome substitution, a method used in conjunction with aneuploid analysis, involves transferring chromosomes from one strain to another. This process is complex and requires careful handling to ensure the successful integration and expression of the transferred chromosomes.
- Potential for Error: Errors in chromosome substitution can lead to incomplete or inaccurate genetic maps, impacting the reliability of subsequent research findings.
Autopolyploidy
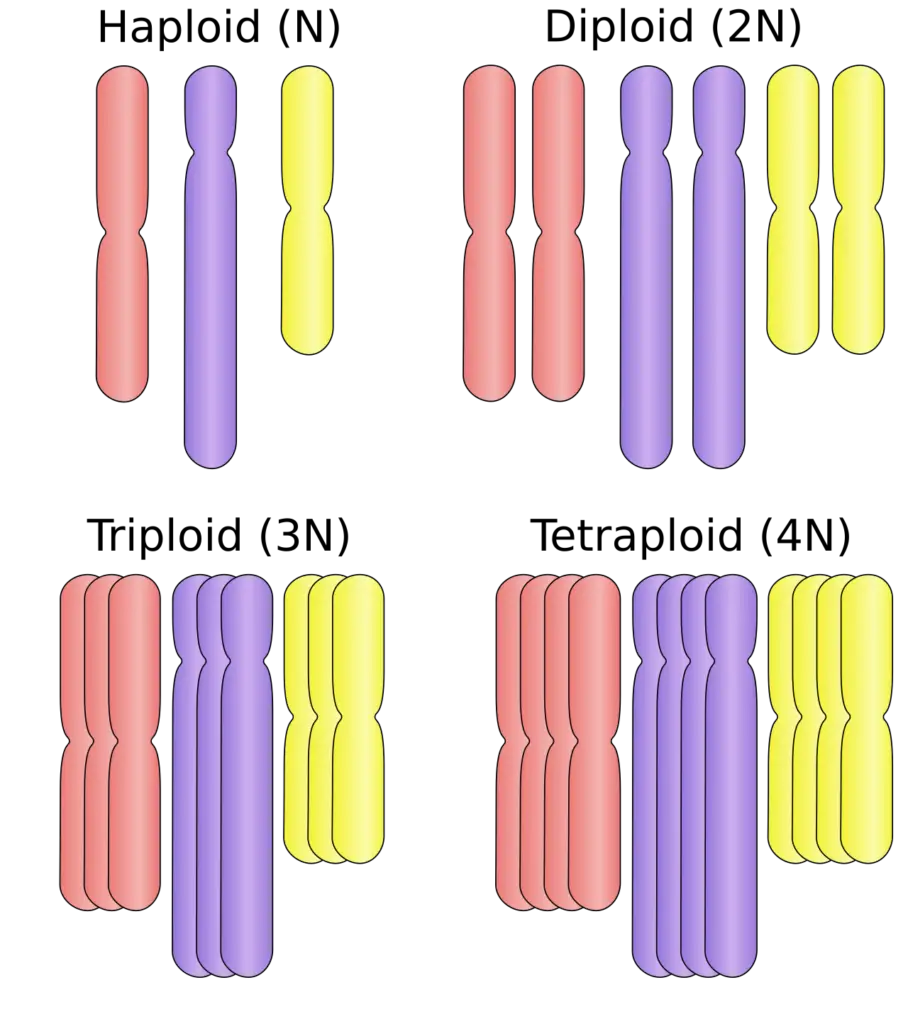
Autopolyploidy refers to the condition where an organism possesses multiple sets of chromosomes derived from a single species. This phenomenon encompasses various levels of ploidy, including monoploidy, triploidy, tetraploidy, and higher ploidy levels. While monoploidy and haploidy are not technically considered forms of polyploidy, they are discussed in the context of autopolyploidy due to their specialized applications in crop improvement. The following points provide a detailed overview of autopolyploidy and its forms:
- Monoploidy and Haploidy:
- Monoploidy involves having a single set of chromosomes (n) and is not traditionally classified as polyploidy. However, it plays a significant role in genetic studies and plant breeding.
- Haploidy refers to organisms with a single set of chromosomes (1n), commonly used to develop homozygous lines in plant breeding.
- Triploidy:
- Definition: Triploidy is characterized by having three sets of chromosomes (3n). It often results from the fusion of a diploid gamete with a tetraploid gamete or through other mechanisms of chromosome doubling.
- Characteristics: Triploids are typically sterile due to irregularities during meiosis, leading to an uneven distribution of chromosomes. This sterility is advantageous in certain crops, such as seedless fruits.
- Tetraploidy:
- Definition: Tetraploidy involves four sets of chromosomes (4n) and can arise through chromosome doubling of a diploid organism or from the fusion of two diploid gametes.
- Applications: Tetraploids often exhibit increased vigor, larger size, and better stress tolerance compared to diploids. This makes them valuable in agricultural breeding programs, particularly in crops like wheat and potatoes.
- Higher Levels of Ploidy:
- Definition: Higher levels of ploidy, such as hexaploidy (6n) and octaploidy (8n), involve even more sets of chromosomes. These levels of ploidy are less common but can occur naturally or be induced artificially.
- Characteristics: Higher ploidy levels may lead to increased robustness and adaptability of plants. However, the complexity of chromosome pairing and segregation can pose challenges for breeding and genetic studies.
- Production of Autopolyploids:
- Direct Production: Autopolyploids can be produced directly through the use of colchicine or other chemical agents that inhibit chromosome segregation during cell division, leading to chromosome doubling.
- Indirect Production: Indirect methods involve the hybridization of species followed by chromosome doubling to restore fertility and stability in the resulting polyploid organism.
- Applications in Crop Improvement:
- Enhanced Traits: Autopolyploids often exhibit desirable traits such as increased size, yield, and stress resistance. These characteristics are utilized in breeding programs to develop improved crop varieties.
- Genetic Stability: Although polyploidy can lead to genetic instability, careful management and selection can harness the benefits of increased chromosome numbers for crop improvement.
Origin and Production of Doubled Chromosome Numbers
Doubling of chromosome numbers can arise through various mechanisms, each with distinct processes and implications. The primary methods for inducing doubled chromosome numbers include spontaneous events, physical treatments, in vitro regeneration, colchicine application, and other chemical agents. Below is a detailed exploration of these methods:
- Spontaneous Chromosome Doubling:
- Description: Occasionally, chromosome doubling can occur spontaneously in somatic tissues or during the formation of unreduced gametes. Such spontaneous events are relatively rare but contribute to genetic variability.
- Mechanism: Unreduced gametes, which contain the diploid chromosome number instead of the usual haploid, can arise due to incomplete meiosis or other genetic anomalies. Certain genes promoting asynapsis or desynapsis may increase the frequency of unreduced gametes.
- Production of Adventitious Buds:
- Description: In some plant species, decapitation or removal of the apical meristem can lead to the formation of callus tissue at the cut site. This callus can occasionally contain polyploid cells.
- Process: Regeneration from such callus tissue can yield adventitious buds, some of which may be polyploid. For example, in Solanaceae plants, up to 36% of regenerated buds have been observed to be tetraploid.
- Enhancement: The application of 1% indole-3-acetic acid (IAA) to the cut end can enhance callus development, potentially increasing the frequency of polyploid buds.
- Physical Agents:
- Description: Physical treatments such as heat, cold, centrifugation, and irradiation can induce chromosome doubling.
- Examples:
- Cold Treatment: Exposure to low temperatures can induce tetraploidy, as seen in Datura plants.
- Heat Treatment: Application of temperatures between 38-45°C during the first division of zygotes in maize results in tetraploid progeny at a frequency of 2-5%. Heat treatments have also been effective in barley, wheat, and rye.
- Irradiation: X-ray or gamma-ray irradiation can cause chromosomal alterations, although its effectiveness in inducing polyploidy is generally lower compared to other methods.
- Regeneration in Vitro:
- Description: Polyploidy often arises during in vitro culture processes. Cells cultured from callus or suspension cultures may exhibit increased ploidy levels.
- Examples: Plants regenerated from callus cultures of species such as Nicotiana, Datura, and rice frequently exhibit various ploidy levels, including polyploidy.
- Colchicine Treatment:
- Description: Colchicine is a widely used chemical for inducing chromosome doubling. It functions by inhibiting spindle formation during cell division, leading to the retention of all chromatids in the nucleus.
- Chemical Properties: Colchicine is derived from the autumn crocus (Colchicum autumnale) and is soluble in alcohol and chloroform but less soluble in hot water. It is crucial to use freshly prepared solutions due to its instability.
- Application Methods:
- Seed Treatment: Seeds can be soaked in colchicine solutions (0.001 to 1%) for 1 to 10 days, with 0.2% being common.
- Seedling Treatment: Germinating seeds may be inverted to expose only the young shoots to colchicine, with treatment durations ranging from 3 to 24 hours.
- Shoot Tip Treatment: Growing shoot apices can be treated with colchicine (0.1 to 1%) using brushes or droppers. Repeated treatments are often necessary. Alternatively, colchicine can be mixed with lanolin paste and applied to the shoot apex.
- Woody Plants: In these plants, colchicine concentrations of around 1% are used for shoot bud treatment.
- Effects and Challenges: Colchicine treatment may lead to variable outcomes, including unintended haploidy or mutations. The effectiveness of colchicine can be influenced by genotype, environmental conditions, and nutritional factors.
- Other Chemical Agents:
- Description: Various other chemicals, such as acenaphthene, 8-hydroxyquinoline, and nitrous oxide, can also induce polyploidy. However, their effectiveness is generally lower compared to colchicine.
- Applications: These chemicals are used less frequently and are not as well-established in inducing polyploidy as colchicine.
Morphological and Cytological Features of Autopolyploids
Autopolyploids exhibit a range of morphological and cytological characteristics that distinguish them from their diploid counterparts. These features are influenced by the level of ploidy and can vary between different species. The following outlines the primary morphological and cytological traits observed in autopolyploids.
Morphological Features
- Cell Size and Stomata:
- Increased Cell Size: Autopolyploids typically have larger cells compared to diploids. This enlargement is evident in various cell types, including guard cells of stomata.
- Stomatal Density: The number of stomata per unit area is usually lower in polyploids than in diploids, partly due to the larger size of each guard cell.
- Pollen Grains:
- Size Increase: Pollen grains of polyploids are generally larger than those of diploids. This increase in size can affect pollen viability and fertilization.
- Growth and Flowering:
- Growth Rate: Polyploids often exhibit slower growth rates compared to diploids. They may also show delayed flowering, which can impact their overall productivity.
- Leaf and Flower Size:
- Leaf and Flower Enlargement: Polyploids usually have larger and thicker leaves, as well as larger flowers and fruits. However, these structures may occur in lower numbers compared to diploids.
- Fertility:
- Reduced Fertility: Polyploids often face reduced fertility due to irregularities during meiosis and genotypic imbalances. These factors lead to physiological disturbances that impact reproductive success.
- Vigor and Growth:
- Variable Vigor: While autopolyploidy can lead to increased general vigor and enhanced vegetative growth in some cases, there are instances where polyploids may be smaller and weaker than their diploid counterparts.
- Optimum Ploidy Levels:
- Species-Specific Optimum Levels: Different species have varying levels of optimum ploidy. For instance, sugarbeet (Beta vulgaris) is most productive at a triploid level, while timothy grass (Phleum pratense) exhibits optimal growth at higher ploidy levels, ranging from 8x to 10x.
- Increased Biomass:
- Higher Biomass: Autopolyploids generally possess increased biomass and better performance in terms of total yield, as seen in crops like tetraploid turnips (Brassica rapa) and cabbage (B. oleracea).
Cytological Features
- Monoploids:
- Chromosome Behavior: In monoploids, chromosomes do not pair during meiosis, resulting in random distribution at anaphase I. This typically leads to almost complete sterility, with some functional gametes producing progeny with 2n chromosomes.
- Triploids:
- Chromosome Pairing: Triploids exhibit variable numbers of trivalents, bivalents, and univalents during meiosis. The irregular separation of chromosomes and distribution of univalents at anaphase I lead to a range of aneuploid gametes, resulting in progeny with diverse chromosome numbers, including trisomics and double trisomics.
- Fertility: Triploids are often highly sterile due to the uneven distribution of chromosomes.
- Tetraploids:
- Chromosome Pairing: Tetraploids display quadrivalents, trivalents, bivalents, and univalents at metaphase I of meiosis. Fertility in tetraploids varies by species but is generally lower than in diploids.
- Fertility Improvement: Despite initial fertility challenges, selection can improve fertility in tetraploids. Successful examples include maize (Zea mays), Petkus rye (Secale cereale), and rice (Oryza sativa). In some cases, such as tetraploid Petkus rye, fertility increased from 60% to 75% over six years of selection.
Segregation in Autotetraploids
The segregation of chromosomes in autotetraploids presents a complex scenario compared to diploids due to the presence of four sets of homologous chromosomes. Understanding this process requires examining how different types of alleles segregate during meiosis and the effects of chromosomal arrangements and crossing over. The following provides a detailed overview of segregation in autotetraploids, with a focus on simplex configurations and associated outcomes.
Segregation Mechanisms
- Types of Allelic Configurations:
- Simplex (Aaaa): Contains one dominant allele and three recessive alleles.
- Duplex (AAaa): Contains two dominant and two recessive alleles.
- Triplex (AAAa): Contains three dominant and one recessive allele.
- Quadruplex (AAAA): Contains four dominant alleles.
- Nulliplex (aaaa): Contains four recessive alleles.
- Random Chromosome Segregation:
- Definition: Occurs when the gene is close to the centromere and no crossing over happens between the centromere and the gene.
- Mechanism: Both sister chromatids of each chromosome move to the same pole at anaphase I. At anaphase II, chromatids carrying dominant alleles will separate and move to opposite poles, but since the dominant alleles do not pair, gametes with dominant alleles are not produced.
- Outcome: This results in two types of gametes, Aa and aa, in a 1:1 ratio. When self-pollinating a simplex individual, the resulting progeny will have three genotypes—AAaa, Aaaa, and aaaa—in a 1:2:1 ratio, with a phenotypic ratio of 3:1 if the dominant allele A produces a dominant phenotype.
- Random Chromatid Segregation:
- Definition: Occurs when crossing over between the centromere and the gene happens, resulting in the attachment of dominant alleles to different chromosomes.
- Mechanism: During crossing over, the two sister chromatids carrying the dominant allele may become attached to different centromeres. At anaphase I, these chromosomes might move to the same pole, and at anaphase II, dominant alleles can end up in the same gamete, leading to AA gametes.
- Outcome: This results in three types of gametes: AA, Aa, and aa, in a ratio of 1:2:1. Selfing a simplex individual under these conditions yields a distribution of progeny that reflects the impact of crossing over, with expected frequencies for various genotypes including quadruplex, triplex, duplex, simplex, and nulliplex.
- Assumptions and Real-World Observations:
- Assumptions:
- Quadrivalents (four-chromosome groupings) are formed regularly.
- Complete or no linkage occurs between the centromere and the gene.
- Reality: These assumptions are often not fully met. Regular formation of quadrivalents and complete linkage are rare, leading to actual gametic and zygotic frequencies being intermediate between the extremes of random chromosome and random chromatid segregation.
- Assumptions:
Role of Autopolyploidy in Evolution
Autopolyploidy plays a notable, albeit selective, role in the evolution of plant species. It has been instrumental in the development of several important crops and has contributed to the diversity of plant forms. Here is a detailed examination of its role in evolution:
- Impact on Crop Species:
- Successful Autopolyploids: Many contemporary crop species are autotetraploids (4x), which have demonstrated significant agricultural value. Examples include:
- Potato (Solanum tuberosum): Cultivated potato has a somatic chromosome number of 48, whereas its wild relatives typically have 24 chromosomes.
- Coffee (Coffea arabica): Cultivated coffee is tetraploid with 44 chromosomes, compared to its wild relatives with varying chromosome numbers.
- Alfalfa (Medicago sativa): A key forage crop, cultivated alfalfa is a tetraploid with 32 chromosomes.
- Peanut (Arachis hypogaea): Another tetraploid, cultivated peanuts have 40 chromosomes, whereas related species exhibit different chromosomal counts.
- Banana (Musa sapientum): Cultivated banana is triploid with 33 chromosomes, distinct from its wild relatives.
- Sweetpotato (Ipomoea batatas): This crop is hexaploid with 90 chromosomes, while its wild relatives have fewer chromosomes.
- Successful Autopolyploids: Many contemporary crop species are autotetraploids (4x), which have demonstrated significant agricultural value. Examples include:
- Role in Crop Improvement:
- Vigor and Yield: Autopolyploidy can enhance certain desirable traits such as increased vigor, size, and productivity. For instance, autotetraploids often exhibit improved growth characteristics compared to their diploid counterparts.
- Genetic Variation: The genetic variability introduced by polyploidy can provide a basis for selection and breeding, leading to the development of new cultivars with improved traits.
- Evolutionary Success:
- Adaptation and Survival: Autopolyploid plants have demonstrated adaptability to various environmental conditions, contributing to their success and prevalence in agriculture.
- Diversity in Forage and Ornamentals: Many forage grasses and ornamental plants are likely autopolyploids, indicating their role in plant diversity and ecosystem functioning.
- Limitations and Considerations:
- Chromosomal Complexity: The complexity of chromosomal arrangements in autopolyploids, such as quadrivalents and trivalents during meiosis, can lead to challenges in fertility and reproduction.
- Speciation: While autopolyploidy contributes to genetic diversity, its role in speciation is more pronounced in some plant families than others.
Application of Autopolyploidy in Crop Improvement
The applications of autopolyploidy in crop improvement are diverse, with specific advantages and challenges associated with different ploidy levels. Here is an organized summary of these applications:
- Monoploids and Haploids:
- Development of Homozygous Lines:
- Monoploids, which are less robust than diploids, offer unique opportunities in crop improvement by facilitating the creation of homozygous diploid lines through chromosome doubling. This method significantly reduces the time and effort required to isolate inbreds and pure lines.
- Isolation of Mutants:
- Monoploids can be used to isolate mutants since recessive alleles, which may not be expressed in diploids, are visible due to the single dose of the gene in somatic tissues. Doubling the chromosome number of these mutants can produce homozygous mutant lines efficiently.
- Increased Selection Efficiency:
- Selection based on haploids or haploid-derived diploids can be more efficient than that based on diploid plants. This is due to the higher frequency of desirable gametes compared to desirable zygotes.
- Breeding Ease in Autotetraploids:
- In autotetraploids like potato, breeding is often easier at the haploid (2x) level compared to the tetraploid (4x) level. This approach simplifies the breeding process and accelerates the development of new varieties.
- Development of Homozygous Lines:
- Triploids:
- Seedless Varieties:
- Triploids, typically produced by hybridizing tetraploid and diploid strains, are generally sterile but are valuable for producing seedless varieties. For example, seedless watermelons are produced by crossing tetraploid (4x) females with diploid (2x) males. These triploid plants do not produce true seeds, which is advantageous for commercial fruit production.
- Enhanced Vigour:
- In some species, such as sugarbeets, triploids can exhibit increased vigor compared to diploids. Triploid sugarbeets, for instance, produce larger roots and higher sugar yields per unit area than diploids.
- Seedless Varieties:
- Tetraploids:
- Improved Quality and Breeding:
- Autotetraploids, which have four sets of chromosomes, have been extensively used to improve crop quality and facilitate breeding. They can enhance traits such as size, vigor, and resistance to pests and diseases.
- Overcoming Self-Incompatibility:
- Tetraploidy can overcome self-incompatibility issues in certain crops. For example, some genotypes of tobacco and white clover exhibit improved compatibility at the tetraploid level.
- Successful Distant Crosses:
- Autotetraploids can sometimes enable successful hybridization between species that are incompatible at the diploid level. For example, certain tetraploid Solanum species can hybridize with S. tuberosum, whereas diploids cannot.
- Forage Crops:
- Tetraploid varieties of forage crops, such as red clover and ryegrass, have shown improved vigor, digestibility, and resistance to pests compared to diploid varieties. Tetraploid turnips and cabbages also exhibit larger size but have higher water content, which can affect their commercial appeal.
- Improved Quality and Breeding:
- Challenges and Limitations:
- Sterility and Genetic Instability:
- Autotetraploids often face challenges related to sterility and genetic instability. These issues can impede seed production and make it difficult to achieve consistent breeding results.
- Success Rates and Applications:
- Autopolyploidy tends to be more successful in species with lower chromosome numbers and in cross-pollinating species rather than self-pollinating ones. Additionally, crops grown for vegetative parts are generally more likely to benefit from polyploidy compared to those grown primarily for seeds.
- Sterility and Genetic Instability:
Limitations of Autopolyploidy
Autopolyploidy, while offering various advantages for crop improvement, also presents several significant limitations that affect its practical application. The following points outline these limitations in detail:
- Increased Water Content and Reduced Dry Matter:
- High Water Content:
- Autopolyploids often exhibit larger sizes compared to their diploid counterparts. However, this increased size is frequently accompanied by higher water content. For crops grown for their vegetative parts, such as turnips (Brassica rapa) and cabbages (Brassica oleracea), this results in a discrepancy where the larger autopolyploids may not produce more dry matter than diploids. Despite higher fresh weights, these polyploids may be comparable or even inferior in terms of dry matter production.
- High Water Content:
- Seed Sterility and Poor Seed Set:
- Low Fertility:
- Autopolyploids, particularly autotetraploids, often suffer from high sterility rates. This sterility leads to poor seed set, which is a critical issue for crops grown primarily for their seeds. Although autotetraploids may have larger seeds, this does not necessarily translate to increased seed yield per unit area.
- Complex Segregation:
- The fertility of autotetraploids can be enhanced through hybridization and selective breeding. Nevertheless, the complex segregation patterns associated with autopolyploids result in slow progress. Achieving acceptable fertility levels can require many years of breeding and selection.
- Low Fertility:
- Challenges in Maintenance and Genetic Stability:
- Clonal Propagation:
- Monoploids and triploids cannot be maintained through traditional sexual reproduction and must be propagated clonally. This limits their practical use in many agricultural settings.
- Genetic Instability:
- The genetic stability of autopolyploids, particularly triploids and autotetraploids, can be problematic. Their progeny often display variability in chromosome number, which complicates maintenance and seed production. This genetic instability presents significant challenges for the consistent production of high-quality crops.
- Clonal Propagation:
- Unfavorable Traits in New Polyploids:
- Desirable Features:
- Newly created polyploids (raw polyploids) frequently exhibit undesirable traits. For instance, they may have weaker stem strength or irregular fruit sizes, as seen in grapes and watermelons, respectively. These inherent defects often prevent immediate use in commercial production.
- Need for Further Improvement:
- Significant improvement through hybridization and selection is generally required to eliminate these defects and enhance the overall performance of new polyploid varieties. This process can be time-consuming and may not always result in successful outcomes.
- Desirable Features:
Allopolyploidy
Allopolyploidy refers to the process of hybridizing two different species to produce a polyploid organism with multiple sets of chromosomes from both parent species. This form of polyploidy has garnered significant interest due to its potential in creating new plant species and improving crop varieties. The following points outline the key aspects of allopolyploidy:
- Definition and Formation:
- Hybridization:
- Allopolyploidy arises from the interspecific hybridization between two distinct species. The resulting hybrid inherits chromosome sets from both parent species.
- Chromosome Doubling:
- After the initial hybridization, chromosome doubling often occurs to stabilize the hybrid’s genome. This process results in an organism with a complete set of chromosomes from each parent, leading to polyploidy.
- Hybridization:
- Applications and Successes:
- Triticale:
- A notable example of successful allopolyploidy is Triticale, a hybrid of wheat (Triticum) and rye (Secale cereale). This crop has been developed for its high yield and adaptability, showing the practical benefits of allopolyploidy in agriculture.
- Raphanobrassica:
- Another example is Raphanobrassica, a hybrid between radish (Raphanus sativus) and cabbage (Brassica oleracea). This allopolyploid demonstrates the potential of creating new crops with desired traits through hybridization.
- Forage Grasses:
- Certain allopolyploids in forage grasses have shown promise, highlighting their potential for improving forage quality and yield.
- Triticale:
- Advantages:
- Increased Genetic Variation:
- Allopolyploids benefit from genetic variation introduced from both parent species. This variation can lead to new traits and enhanced adaptability.
- Hybrid Vigor:
- Many allopolyploids exhibit hybrid vigor (heterosis), which can result in improved growth rates, yield, and resistance to diseases compared to their parent species.
- Increased Genetic Variation:
- Challenges and Considerations:
- Complexity in Breeding:
- Breeding allopolyploid plants can be complex due to the need for maintaining stability of multiple sets of chromosomes. This complexity requires careful management and selective breeding.
- Fertility Issues:
- Some allopolyploids may experience fertility issues, affecting their ability to reproduce and establish new populations. This can limit their practical applications in agriculture.
- Complexity in Breeding:
Origin and Production of Allopolyploids
Allopolyploids arise from hybridization between different species, which subsequently undergo chromosome doubling. This section outlines the mechanisms through which allopolyploids originate and are produced.
Origin of Allopolyploids
- Hybridization:
- Initial Crosses:
- Allopolyploids typically originate from the hybridization of two distinct species, which may belong to the same genus or different genera. These hybrids, termed F1 hybrids, possess chromosomes from both parent species.
- Chromosome Doubling:
- Chromosome doubling is crucial for stabilizing the hybrid’s genome. This can occur through several mechanisms:
- Somatic Doubling: Irregular mitotic cell division in somatic tissues can result in chromosome doubling. This may happen in the apical meristem or axillary buds, producing allopolyploid sectors or branches.
- Unreduced Gametes: Irregular meiosis can produce unreduced gametes with a full set of chromosomes. Fusion of these gametes can yield allopolyploid progeny.
- Chromosome doubling is crucial for stabilizing the hybrid’s genome. This can occur through several mechanisms:
- Initial Crosses:
- Examples of Natural Occurrence:
- Primula verticillata x P. floribunda:
- In this cross, a fertile branch with double the chromosome number of the F1 hybrid was observed. Progeny from this branch were identified as allotetraploid (Primula kewensis).
- Raphanobrassica:
- This allopolyploid resulted from a cross between cabbage (Brassica oleracea) and radish (Raphanus sativus). Despite the F1 hybrid being largely sterile, occasional unreduced gametes led to the formation of the allotetraploid.
- Primula verticillata x P. floribunda:
Production of Allopolyploids
- Experimental Techniques:
- Chromosome Doubling:
- To artificially produce allopolyploids, researchers often double the chromosome number of distant hybrids. This is typically achieved using colchicine, a chemical that inhibits spindle formation during cell division.
- Synthetic Allopolyploids:
- Allopolyploids created through artificial means are known as synthetic allopolyploids. This involves:
- Creating the F1 Hybrid:
- The first step is to produce a distant hybrid from two distinct plant species.
- Doubling the Chromosome Number:
- The second step is to induce chromosome doubling in the hybrid using colchicine or other agents.
- Creating the F1 Hybrid:
- Allopolyploids created through artificial means are known as synthetic allopolyploids. This involves:
- In Vitro Techniques:
- Modern techniques, including in vitro methods, have facilitated the production of distant crosses that were previously challenging. While colchicine is commonly used, other agents may be employed if a species shows poor response to colchicine.
- Chromosome Doubling:
- Challenges and Considerations:
- Species Response:
- Most plant species respond well to colchicine, but some may exhibit poor responsiveness. In such cases, alternative agents for chromosome doubling might be necessary.
- Species Response:
Morphological and Cytological Features of Allopolyploids
Allopolyploids exhibit a unique set of morphological and cytological features resulting from their hybrid origins. Understanding these features involves examining both the visible characteristics and the chromosomal configurations of these plants.
Morphological Features
- Combination of Parental Traits:
- General Characteristics:
- Allopolyploids often combine traits from their parent species. However, predicting the exact combination of these traits can be challenging.
- Specific Examples:
- Raphanobrassica:
- The goal was to merge the root of radish (Raphanus sativus) and the leaves of cabbage (Brassica oleracea). The resulting plant exhibited radish-like leaves and cabbage-like roots, highlighting the unpredictable nature of trait combination in allopolyploids.
- Triticale:
- This allopolyploid successfully combined the hardiness of rye (Secale cereale) and the high yield of wheat (Triticum spp.), demonstrating a more predictable trait combination.
- Raphanobrassica:
- General Characteristics:
- Vigor and Adaptability:
- General Observations:
- Allopolyploids generally show increased vigor compared to diploids. This enhanced vigor is often associated with improved adaptability.
- Ecological Adaptation:
- While allopolyploids are frequently more adaptable than their diploid ancestors, they rarely displace these ancestors from their ecological niches. Evolutionary success of allopolyploids often involves colonizing new ecological areas.
- General Observations:
- Apomixis:
- Definition and Occurrence:
- Many allopolyploids are apomictic, meaning they reproduce asexually without fertilization. This is common in certain grasses, such as Poa, and in other plant groups like Taraxacum and Rubus.
- Function:
- Apomixis provides a reproductive advantage in situations where hybrid sterility is a problem, allowing these plants to maintain genetic stability.
- Definition and Occurrence:
Cytological Features
- Chromosomal Composition:
- Homoeologous Chromosomes:
- Chromosomes in allopolyploids often include homoeologous chromosomes—those that are similar but not identical to each other. These result from hybridization between related species.
- Amphidiploids:
- After chromosome doubling, allopolyploids possess two sets of chromosomes from each parent species, leading to a diploid-like chromosomal arrangement known as amphidiploid or amphiploid.
- Homoeologous Chromosomes:
- Chromosome Pairing:
- Divergence and Pairing:
- When the two genomes in an amphidiploid are divergent, pairing is typically regular, with only bivalents forming at metaphase I. In contrast, if the genomes are more similar (homoeologous), quadrivalents may form, leading to variable chromosome pairing.
- Fertility Implications:
- Variable pairing can affect fertility. Amphidiploids with many quadrivalents may be partially fertile but exhibit genetic instability.
- Divergence and Pairing:
- Diploidization:
- Process and Outcome:
- Through hybridization and selection, the fertility and chromosomal behavior of allopolyploids can be improved. This process, known as diploidization, enhances bivalent formation, making the allopolyploid function more like a diploid in terms of chromosome behavior.
- Genetic Systems:
- Natural allopolyploids often develop efficient genetic systems to ensure regular bivalent formation. For example, Triticum aestivum (wheat) has a genetic mechanism that suppresses homoeologous pairing, contributing to its stability.
- Process and Outcome:
Role of Allopolyploidy in Evolution
Allopolyploidy plays a significant role in the evolution of plant species, contributing to both the diversity and adaptability of crops. The evolutionary importance of allopolyploids can be understood through their formation, characteristics, and the impact they have had on agriculture and natural ecosystems.
Evolutionary Significance
- Prevalence and Success:
- Widespread Occurrence:
- Allopolyploids are prevalent among angiosperms, with estimates suggesting that approximately one-third of these flowering plants are polyploids. Most of these are allopolyploids, indicating their major role in plant evolution.
- Crop Success:
- Allopolyploids have been particularly successful as crop species. This success is reflected in their widespread use and the variety of crops that are allopolyploid, including wheat, cotton, and Brassica species.
- Widespread Occurrence:
- Historical Insights:
- Early Theories:
- In 1917, Winge proposed that allopolyploidy was crucial for evolutionary processes. Subsequent research has supported this idea, showing that allopolyploidy contributes significantly to plant diversity and adaptation.
- Tracing Evolutionary History:
- The evolutionary history of many allopolyploid crops has been traced back to their diploid ancestors. This process often involves studying chromosome pairing between diploids and allopolyploids to identify parental species.
- Early Theories:
- Chromosomal Evidence:
- Chromosome Pairing:
- The pairing of chromosomes between diploid and allopolyploid species provides clues to the evolutionary relationships. Homology between chromosomes of the diploid and allopolyploid species suggests that the diploid species are likely ancestors of the allopolyploid.
- Synthetic Allopolyploids:
- Synthesizing allopolyploids from known diploid species can help confirm the parental origins. For example, the synthetic allopolyploid Brassica napus closely resembles its natural counterpart, indicating a successful reconstruction of evolutionary relationships.
- Chromosome Pairing:
Case Studies
- Wheat (Triticum spp.):
- Genome Composition:
- Wheat is a well-known example of an allopolyploid with complex evolutionary history. The A, B, and D genomes of Triticum aestivum have been investigated through chromosome pairing studies. These studies suggest that the A genome is similar to that in tetraploid and hexaploid wheats, while the B genome is similar to that in tetraploid wheats.
- Challenges in Identification:
- Despite extensive research, the exact diploid species contributing to the A, B, and D genomes are still debated. This uncertainty highlights the complexity of tracing evolutionary origins in polyploid species.
- Genome Composition:
- Tobacco (Nicotiana tabacum):
- Hybrid Origins:
- Nicotiana tabacum is believed to be an amphidiploid resulting from the cross between Nicotiana sylvestris and Nicotiana tomentosa. Chromosome pairing studies confirm that N. tabacum shares homology with these diploid species.
- Evolutionary Differentiation:
- Over time, N. tabacum has undergone significant differentiation, including gene mutations and the loss of some duplicated segments, which have contributed to its current characteristics.
- Hybrid Origins:
- Cotton (Gossypium spp.):
- Origins and Evolution:
- Gossypium hirsutum is thought to have originated from a cross between Gossypium arboreum and Gossypium thurberi, followed by chromosome doubling. Alternative hypotheses suggest origins involving Gossypium herbaceum var. africanum and Gossypium raimondii.
- Chromosomal Differences:
- Different species of cotton exhibit varying chromosome sizes, which reflect their evolutionary histories and adaptations.
- Origins and Evolution:
- Brassica Species:
- Amphidiploid Origins:
- The evolution of Brassica species, such as B. napus and B. juncea, has been mapped using the U’s Triangle. This model illustrates how different diploid species, such as B. oleracea and B. campestris, have contributed to the formation of these amphidiploid species.
- Synthetic vs. Natural Allopolyploids:
- Synthetic allopolyploids often resemble natural counterparts, confirming the role of specific diploid species in their evolution. However, the resemblance is not always perfect, indicating the complexities of allopolyploid evolution.
- Amphidiploid Origins:
Challenges in Determining Parentage
- Extinction of Parental Species:
- Loss of Ancestral Forms:
- Some diploid parental species may no longer exist, making it difficult to trace the exact origins of allopolyploids.
- Loss of Ancestral Forms:
- Genetic Divergence:
- Changes Over Time:
- Both diploid and allopolyploid species may have undergone significant genetic changes since their hybridization, complicating the identification of parental species.
- Changes Over Time:
- Evolutionary Changes in Allopolyploids:
- Differentiation:
- Allopolyploids themselves may have evolved considerably from their original forms, further obscuring their evolutionary history.
- Differentiation:
Applications of Allopolyploids in Crop Improvement
Allopolyploids, formed through hybridization between different species followed by chromosome doubling, have several significant applications in crop improvement. Their role extends across bridging species for gene transfer, creating new crop species, and broadening the genetic base of existing crops. Each application leverages the unique attributes of allopolyploids to address specific agricultural challenges.
1. Utilization as Bridging Species
- Role in Gene Transfer:
- Introduction of Novel Traits:
- Allopolyploids can act as intermediaries to transfer desirable traits from one species to another. This is particularly useful when hybrids between a cultivated species and a wild species are sterile.
- Process:
- When a hybrid between a recipient crop species and a donor wild species is sterile, an amphidiploid can be used to bridge the gap. The amphidiploid, which is fertile, contains genomes from both species. By backcrossing this amphidiploid with the recipient species, it is possible to produce progeny that have the chromosome number of the recipient species but also carry some genes from the donor species.
- Introduction of Novel Traits:
- Examples:
- Tobacco Mosaic Virus Resistance:
- An amphidiploid like Nicotiana digluta (allohexaploid) can be used to transfer resistance to tobacco mosaic virus from Nicotiana sylvestris to Nicotiana tabacum. This process involves creating a pentaploid that is sufficiently fertile for backcrossing to N. tabacum, leading to resistant plants.
- Cotton and Wheat:
- The transfer of traits from Gossypium thurberi to Gossypium hirsutum and from Haynaldia villosa to Triticum aestivum illustrates the application of amphidiploids in improving these crops. These processes have been successful in incorporating valuable traits into commercially important species.
- Tobacco Mosaic Virus Resistance:
2. Creation of New Crop Species
- Historical Context and Expectations:
- Early Optimism:
- There was initial excitement about the potential of allopolyploidy to create new and superior crop species by mimicking natural processes of polyploid formation.
- Challenges and Realities:
- Despite early hopes, newly synthesized allopolyploids often face challenges. They typically do not outperform existing species because natural allopolyploids have undergone long periods of evolution, refining their traits. Newly synthesized allopolyploids may exhibit low fertility and other issues, which limits their immediate practical applications.
- Early Optimism:
- Successful Examples:
- Triticale:
- The synthetic allopolyploid Triticale, produced by crossing wheat (tetraploid or hexaploid) with rye, has been a notable success. Triticale combines the high yielding capacity of wheat with the hardiness of rye. After extensive research and breeding, triticale now exhibits comparable yields to the best wheat varieties in some regions, such as Canada.
- Other Promising Species:
- Raphanobrassica, Festuca-Lolium hybrids, and interspecific hybrids in Rubus and Jute represent other examples of synthetic allopolyploids with potential, though they also face issues like low fertility and genetic instability.
- Triticale:
3. Widening the Genetic Base of Existing Allopolyploids
- Genetic Improvement:
- Addressing Narrow Genetic Diversity:
- Existing allopolyploids often have limited genetic variability. To enhance their adaptability and yield, new allopolyploid variants can be synthesized. For example, in Brassica napus, which has a narrow genetic base, new allopolyploids are being created by crossing B. campestris with B. oleracea to introduce greater genetic diversity.
- Methodology:
- The process involves complex breeding techniques, including the use of embryo culture to overcome difficulties in crossing autotetraploids. The goal is to produce variants with enhanced traits that can be integrated into commercial breeding programs.
- Addressing Narrow Genetic Diversity:
Limitations of Allopolyploidy
- Predictability and Stability:
- Unpredictable Outcomes:
- The effects of allopolyploidy can be unpredictable. While some allopolyploids may exhibit desirable traits, others may inherit undesirable features from their parental species.
- Defects and Challenges:
- Newly synthesized allopolyploids often suffer from low fertility, cytogenetic and genetic instability, and other issues. These problems require extensive breeding efforts to overcome.
- Resource Intensive:
- The development of new allopolyploid crop species involves considerable time, labor, and resources. Only a small proportion of allopolyploids prove to be commercially viable, making the process of discovery and improvement both costly and lengthy.
- Unpredictable Outcomes:
- https://www.shcollege.ac.in/wp-content/uploads/NAAC_Documents_IV_Cycle/Criterion-II/2.3.2/ppt/Dr_ImaNeerakkal_Polyploidyshortfinal.pdf
- https://ddugu.ac.in/ePathshala_Attachments/[email protected]
- https://www.researchgate.net/profile/Geoffrey-Meru/publication/256199268_Polyploidy_and_its_implications_in_plant_breeding/links/00463522002bc5a024000000/Polyploidy-and-its-implications-in-plant-breeding.pdf
- https://bsmrau.edu.bd/seminar/wp-content/uploads/sites/318/2018/06/Tamanna-seminar.pdf
- https://iastate.pressbooks.pub/cropgenetics/chapter/ploidy-polyploidy-aneuploidy-haploidy-2/
- https://rfna.com.my/archive/1rfna2023/1rfna2023-25-27.pdf