What is chitin?
- Chitin is a natural polysaccharide composed of repeating N-acetylglucosamine units.
- It forms the primary structural component of arthropod exoskeletons (insects, crustaceans) and fungal cell walls.
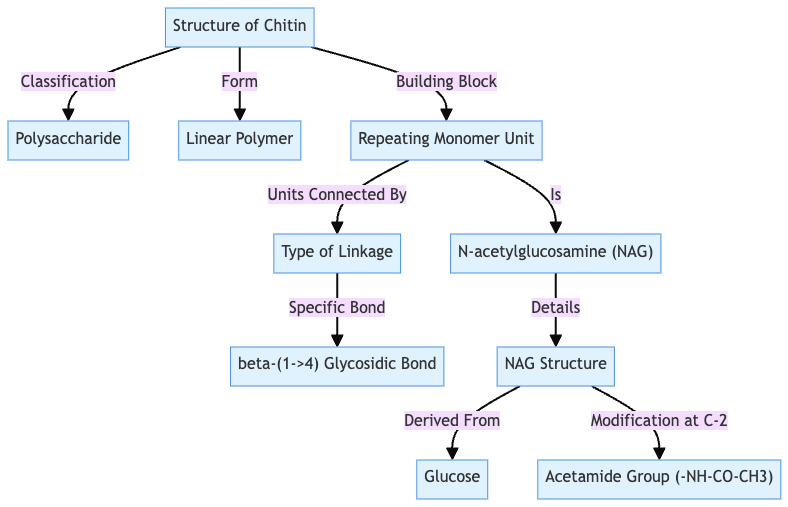
- Its β-1,4-glycosidic bonds create a rigid, insoluble matrix that resists degradation
- Chitin provides mechanical strength, protection against desiccation and microbial attack
- It is biodegradable, biocompatible and used in wound dressings, drug delivery systems and water treatment.
- Industrially, chitin is sourced from shrimp and crab shells and can be deacetylated to produce chitosan.
Structure of chitin
- Chitin is a β(1,4) homopolymer of N-acetyl glucosamine, similar to cellulose.
- The OH group at C2 of each β-D-glucose unit is replaced by an N-acetylamino group.
- Chitin is insoluble in both water and non-polar solvents despite the charges on the acetyl groups.
- Linear chains of N-acetyl-D-glucosamine are linked by β-1,4-glucosidic bonds to form an α-helix.
- The α-helix is stabilized by hydrogen bonding of the N-acetyl side chains.
- In nature, chitin polymers bind extracellularly via intermolecular hydrogen bonds to form crystalline microfibrils.
- Chitin has three forms based on fiber orientation
- α-chitin has antiparallel fibers in an orthorhombic arrangement
- intrasheet hydrogen bonds between amide I and amide II carbonyls
- intersheet hydrogen bonds between CH₂OH side chains and carbonyl groups
- strong hydrogen bonding in the a and b directions; weaker in the c direction
- highest decomposition temperature at 330 °C
- β-chitin has parallel fibers in monoclinic units
- stabilized only by intrasheet hydrogen bonds
- lowest decomposition temperature at 230 °C because of fewer hydrogen bonds
- γ-chitin has alternating parallel and antiparallel fibers
- combines both intersheet and intrasheet hydrogen bonding like α-chitin
- intermediate decomposition temperature at 310 °C
- α-chitin has antiparallel fibers in an orthorhombic arrangement
What are Chitinases?
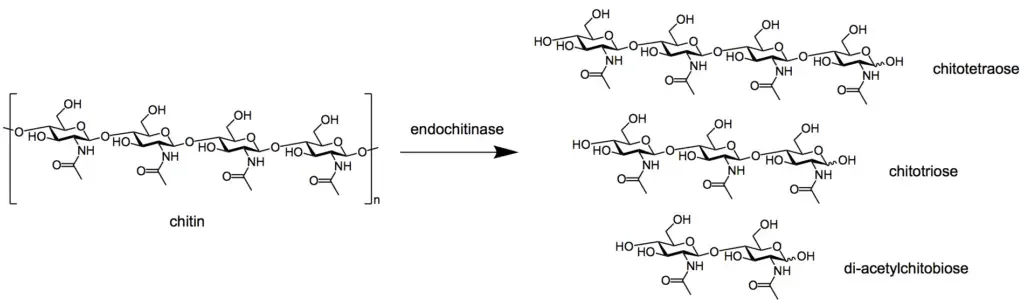
Chitinases are enzymes that catalyze the hydrolysis of the β-1,4-glycosidic bonds in chitin
They belong to the glycosyl hydrolase family with EC number 3.2.1.14
Two main types exist based on cleavage mode
- endochitinases cleave internal bonds within the chitin polymer
- exochitinases remove monomers or dimers from the ends of the chain
They are produced by bacteria, fungi, plants and some animals for nutrition, defense or developmental processes
In plants, chitinases function as pathogenesis-related proteins that degrade fungal cell walls and enhance disease resistance
In microbes, chitinases enable utilization of chitin as a carbon and nitrogen source and support marine and soil nutrient cycling
In mammals, true chitinases and chitinase-like proteins modulate immune responses and have been linked to asthma, allergy and tissue remodeling
Industrial applications include biocontrol of fungal pathogens, conversion of shellfish waste into valuable chito-oligosaccharides, protoplast preparation in fungal research and generation of bioactive compounds
Factors affecting chitin degradation
Temperature
- Higher temperatures generally increase chitin degradation rates by increasing enzyme activity, typically optimal between 30–50 °C; above this range, activity declines or enzymes denature
- Extremophiles like Thermomyces lanuginosus and Pyrococcus chitonophagus have thermostable chitinases active at 45–60 °C and even 120 °C for hyperthermophiles
pH
- Acidic to neutral pH (6–8) generally favors maximum chitinase activity; optimal pH often is around 6–8 depending on the organism
- Extreme pH shifts change chitin chain flexibility and enzyme–substrate interactions
Substrate availability
- Higher chitin concentrations induce chitinolytic microbe and enzyme production
- Presence of easily degradable carbon sources (e.g., glucose) can suppress chitinase production due to catabolite repression
Microbial community composition
- Diversity and abundance of chitin degraders affects degradation rates and pathway efficiency
- Isolated or unique ecosystems select for communities with specific temperature optima
Moisture and aeration
- Adequate moisture and oxygen boosts microbe and enzyme activity
Presence of inhibitors or metal ions
- Certain ions (e.g., Zn²⁺, Cu²⁺) and detergents (e.g., SDS) can inhibit specific chitinases Heavy metals like Ag⁺, Hg²⁺, Fe³⁺ and Cd²⁺ also inhibit N-acetyl-β-D-glucosaminidases
Organic matter and alternative substrates
- High levels of organic matter or alternative carbon sources can shift microbe priorities away from chitin degradation
Incubation time and microbial growth phase
- Chitinase production often peaks after several days (e.g., 5–6 days in Trichoderma)
Which Microorganisms are involved in chitin degradation?
- Chitin degradation is carried out by diverse microorganisms including bacteria, fungi, archaea, protozoa, algae and rotifers
- bacteria—principal agents, especially in soil and aquatic environments
- fungi—significant in soils depending on conditions
- archaea—e.g. Pyrococcus chitonophagus, a hyperthermophile capable of chitinase production
- protozoa, algae and rotifers—capable of chitin breakdown or oligomer processing
- Major bacterial genera involved include
- Streptomyces spp. (e.g. S. lunalinharesii, S. thermoviolaceus)—produce extracellular chitinases, often thermostable.
- Chitinophaga spp. (e.g. C. pendula)—soil bacteria specialized in chitin degradation.
- Isoptericola jiangsuensis—Gram‑positive soil bacterium with chitinolytic activity.
- Marine genera such as Pseudoalteromonas, Motilimonas, Arcobacter, Halarcobacter—active in oceanic chitin turnover.
- Fungal contributors
- Thermophilic fungi like Thermomyces lanuginosus produce thermostable chitinases and colonize compost/heated environments.
- Soil- and plant-associated fungi also participate in chitin degradation depending on pH and temperature conditions.
- Archaeal involvement
- Pyrococcus chitonophagus degrades chitin at high temperatures, producing a 70 kDa chitinase (Chi70) active up to ~120 °C, contributing in hydrothermal environments
- Other microbial groups
- Marine cyanobacteria like Synechococcus possess chitinase genes and can degrade chitin oligomers
- Protozoa, algae, rotifers and even carnivorous plants contribute to chitin breakdown in specific ecosystems
Which Enzymes are involved in the degradation of chitin?
Chitin degradation involves multiple enzymes acting in concert through hydrolytic and oxidative pathways
- Chitinases (glycoside hydrolases EC 3.2.1.14)
- Endochitinases cleave internal β‑1,4 bonds, releasing chitooligosaccharides
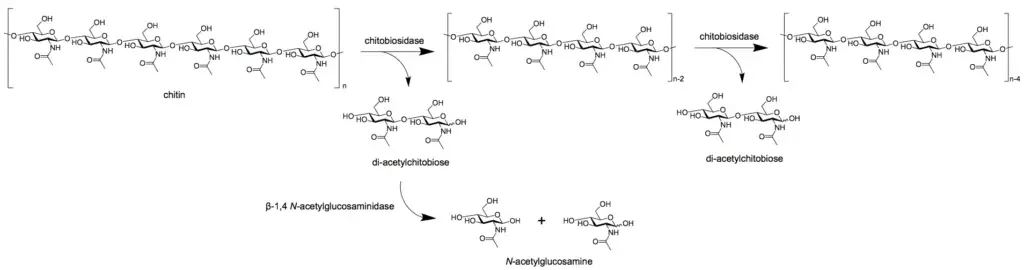
- Exochitinases, including chitobiosidases, act at polymer ends to liberate dimers
- N‑acetyl‑β‑D‑glucosaminidases (NAGases, EC 3.2.1.30/52) hydrolyze chitooligosaccharides into GlcNAc monomers
- Chitosanases
- Similar to chitinases but target partially or fully deacetylated chitin (chitosan); activity depends on degree of acetylation
- Chitin deacetylases (CDAs, EC 3.5.1.41) and chitin oligosaccharide deacetylases (EC 3.5.1.105)
- Remove acetyl groups converting chitin into chitosan and releasing acetate
- Lytic Polysaccharide Monooxygenases (LPMOs, EC 1.14.99.53–56)
- Copper-dependent oxidative enzymes that cleave crystalline chitin by oxidizing C1/C4 carbons, enhancing access for chitinases
- Auxiliary enzymes
- N,N′‑diacetylchitobiose phosphorylase catalyzes phosphorolysis of chitobiose into GlcNAc and GlcNAc‑1‑phosphate, integrating chitin fragments into metabolism
- Supporting enzymes
- Cellulases and lysozymes may exhibit minor chitin-degrading activity, particularly on soluble substrates
- Summary of pathways
- Hydrolytic pathway: chitinases → chitooligosaccharides → NAGases → GlcNAc
- Deacetylation pathway: CDAs convert to chitosan, enabling further degradation
- Oxidative pathway: LPMOs oxidatively disrupt crystalline structure for enhanced degradation
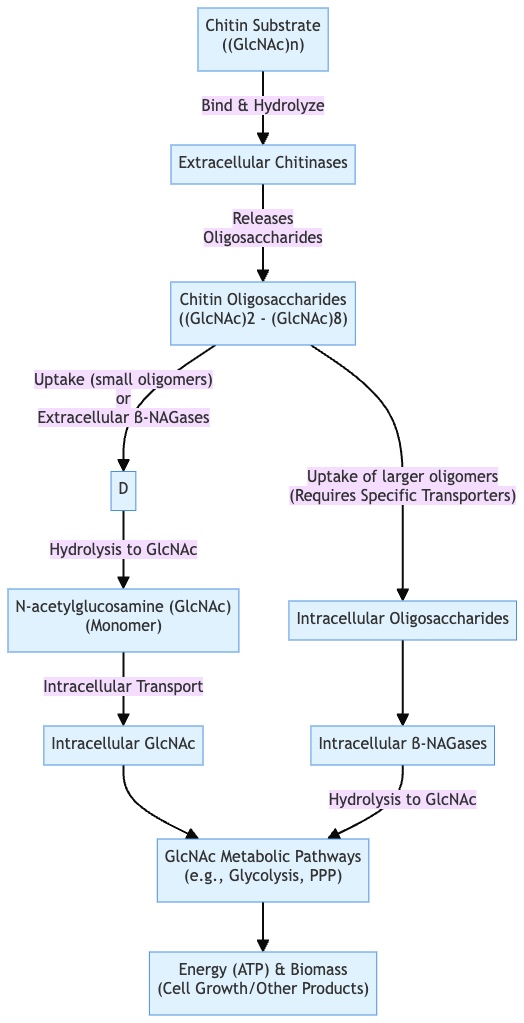
Simple Steps of chitin degradation
These steps outline the coordinated hydrolytic and oxidative pathways microorganisms use to degrade chitin efficiently.
- Depolymerization
- Extracellular chitinases (endo‑ and exo‑types) cleave β‑1,4 bonds in chitin, yielding shorter chitooligosaccharides
- Oxidative cleavage (optional)
- Lytic polysaccharide monooxygenases (LPMOs) oxidize crystalline chitin surfaces, creating nicks and oxidized oligomers (e.g., GlcNAc₁A), which enhance hydrolytic enzyme access
- Oligomer transport
- Chitooligosaccharides are taken up into the periplasm or cytoplasm via specific transport systems (e.g., TBDRs, specific porins)
- Oligomer hydrolysis
- Exochitinases (chitobiosidases) and N‑acetyl‑β‑d‑glucosaminidases (NAGases) break down oligomers into monomeric GlcNAc or oxidized GlcNAc₁A units
- Deacetylation (optional)
- Chitin deacetylases remove acetyl groups, converting chitin or oligomers to chitosan or glucosamine derivatives
- Metabolic assimilation
- GlcNAc enters central metabolism; oxidized monomers (GlcNAc₁A) may be further processed into 2‑keto‑3‑deoxygluconate‑6‑phosphate, acetate, NH₃
Mechanisms of microbial degradation of chitin
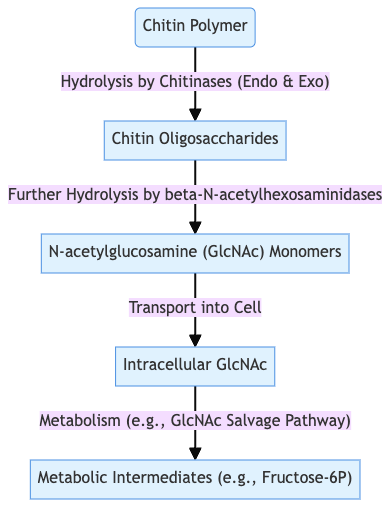
Microorganisms degrade chitin via two main mechanisms: hydrolytic enzymes and oxidative enzymes
Hydrolytic pathway
- Extracellular chitinases (endo‑ and exo‑types) cleave β-1,4 bonds in polymeric chitin, releasing soluble chitooligosaccharides
- These oligomers are transported into the cell via specific transporters
- Intracellular N-acetyl-β-D-glucosaminidases (NAGases) hydrolyze oligomers into N-acetylglucosamine (GlcNAc) monomers
- GlcNAc enters central metabolism, generating energy, acetate, ammonia, and fructose-6-phosphate
Oxidative pathway (LPMO-initiated)
- Lytic polysaccharide monooxygenases (LPMOs) are copper-dependent enzymes that oxidize and nick crystalline chitin, increasing its accessibility to chitinases
- Oxidation generates specific oxidized chitooligosaccharides (e.g., GlcNAc₁A)
- These oligomers are transported into the periplasm or cytoplasm via oxidized-oligosaccharide-specific transporters
- Intracellular enzymes such as OngA hydrolyze oxidized oligomers, further deacetylate and deaminate them
- Products are integrated into metabolic pathways distinct from the hydrolytic route, including deacetylated amino-sugar metabolism.
Synergistic action
- LPMOs and chitinases work synergistically: oxidative nicks by LPMOs enhance chitinase efficiency.
- This synergism is critical for breaking down crystalline chitin in marine and soil environments.
Community processes
- Many bacteria share hydrolytic and oxidative functions in microbial consortia, enhancing degradation through cross-feeding of intermediates
FAQ
What is chitin, and why is it important?
Chitin is a polysaccharide that is found in the exoskeletons of arthropods, as well as in the cell walls of fungi and some algae. It is the second most abundant biopolymer on earth after cellulose, and it plays an important role in the ecology of many ecosystems.
How do microorganisms degrade chitin?
Microorganisms can degrade chitin through two main mechanisms: chitinoclastic and deacetylation. Chitinases are the key enzymes involved in chitinoclastic degradation, while chitosanases and other enzymes are involved in deacetylation.
What are some examples of microorganisms that can degrade chitin?
Many microorganisms can degrade chitin, including bacteria, fungi, and some protozoa. Some examples of chitin-degrading microorganisms include Vibrio cholerae, Serratia marcescens, and Trichoderma reesei.
What are the products of chitin degradation?
The products of chitin degradation can vary depending on the specific microorganisms and conditions involved. However, common products include chitin oligomers, chitosan, and monomeric N-acetylglucosamine.
What is the role of chitin-degrading microorganisms in the ecosystem?
Chitin-degrading microorganisms play an important role in the recycling of organic matter in the ecosystem. They break down chitin-containing waste products, such as exoskeletons and fungal cell walls, into simpler compounds that can be utilized by other organisms.
Can chitin-degrading microorganisms be used for biotechnological applications?
Yes, chitin-degrading microorganisms are of interest for various biotechnological applications, such as the production of chitin oligomers, chitosan, and other value-added compounds.
What factors can affect chitin degradation?
Many factors can influence chitin degradation, including pH, temperature, nutrient availability, oxygen levels, and the activity of other microorganisms in the ecosystem.
How is chitin degradation studied in the laboratory?
Chitin degradation can be studied in the laboratory using various techniques, such as enzyme assays, microbial culture techniques, and molecular biology methods.
What are some challenges associated with chitin degradation?
Chitin degradation can be challenging due to the complexity of the chitin molecule, the presence of other polymers in the ecosystem, and the variability of chitin-degrading microorganisms.
What is the potential impact of chitin degradation on global carbon and nitrogen cycling?
Chitin degradation plays an important role in global carbon and nitrogen cycling, as it contributes to the recycling of organic matter in the ecosystem. However, the exact impact of chitin degradation on these processes is still being studied.
- Gooday, G.W. Physiology of microbial degradation of chitin and chitosan. Biodegradation 1, 177–190 (1990). https://doi.org/10.1007/BF00058835
- Jiang, WX., Li, PY., Chen, XL. et al. A pathway for chitin oxidation in marine bacteria. Nat Commun 13, 5899 (2022). https://doi.org/10.1038/s41467-022-33566-5
- https://communities.springernature.com/posts/a-pathway-for-chitin-oxidation-in-marine-bacteria
- https://www.bohrium.com/paper-details/a-pathway-for-chitin-oxidation-in-marine-bacteria/864962046318346613-10026
- Meunier L, Costa R, Keller-Costa T, Cannella D, Dechamps E, George IF.2024.Selection of marine bacterial consortia efficient at degrading chitin leads to the discovery of new potential chitin degraders. Microbiol Spectr12:e00886-24.https://doi.org/10.1128/spectrum.00886-24
- Elieh-Ali-Komi D, Hamblin MR. Chitin and Chitosan: Production and Application of Versatile Biomedical Nanomaterials. Int J Adv Res (Indore). 2016 Mar;4(3):411-427. Epub 2016 Mar 1. PMID: 27819009; PMCID: PMC5094803.
- Beier, S., & Bertilsson, S. (2013). Bacterial chitin degradation—mechanisms and ecophysiological strategies. Frontiers in Microbiology, 4. doi:10.3389/fmicb.2013.00149
- Beier S, Bertilsson S. Bacterial chitin degradation-mechanisms and ecophysiological strategies. Front Microbiol. 2013 Jun 14;4:149. doi: 10.3389/fmicb.2013.00149. PMID: 23785358; PMCID: PMC3682446.
- https://biologydictionary.net/chitin/
- https://en.wikipedia.org/wiki/N-acetyl-%CE%B2-d-glucosaminidase