What is Introns?
- Introns are non-coding DNA sequences that are present within a gene and are removed through a process called RNA splicing during the maturation of RNA molecules. The term “intron” refers to the intragenic region within a gene, which includes both the DNA sequences within the gene and the corresponding sequences in RNA transcripts.
- Introns are commonly found in the genes of many eukaryotic organisms, as well as in some viruses. They are present in a wide range of genes, including those involved in protein synthesis, ribosomal RNA (rRNA) production, and transfer RNA (tRNA) synthesis. However, introns are not found in prokaryotes.
- The presence and distribution of introns vary among different organisms. For example, protein-coding nuclear genes of most jawed invertebrates have a high prevalence of introns, while they may be rare in some other eukaryotic organisms. Similarly, the mitochondrial genomes of jawed vertebrates typically lack introns, whereas other eukaryotes often contain numerous introns in their mitochondrial DNA.
- There are different types of introns based on their sequence analysis and the mechanisms involved in their removal during RNA splicing. Some common types of introns include spliceosome introns, which are found in nuclear protein-coding genes and are removed by spliceosomes, and tRNA introns, which occur in nuclear and archaeal tRNA genes and are removed by proteins. Additionally, there are self-splicing group I and group II introns, which are removed through RNA catalysis.
- Introns play important roles in gene regulation and protein diversity. Alternative splicing, a controlled molecular mechanism, allows introns to contribute to the production of multiple variant proteins from a single gene in eukaryotic cells. This process greatly enhances the diversity of protein types that can be generated. Introns have also been found to enhance the level of gene expression, and spliced transcripts are reported to be exported from the nucleus to the cytoplasm faster than unspliced transcripts.
- However, the presence of introns in the genome can also be energetically demanding for cells, as they require significant energy for accurate excision and removal at the correct positions with the help of complex spliceosomal machinery.
- In summary, introns are non-coding DNA sequences that are present within genes in eukaryotes. They are removed through RNA splicing and play important roles in gene regulation, protein diversity, and the overall complexity of gene expression in eukaryotic organisms.
Functions of Introns
- Introns, once considered “junk DNA,” have been found to serve important functions in gene regulation and genetic diversity.
- One of the functions of introns is to increase genetic variation. By increasing the length of genes, introns promote crossover and recombination between sister chromosomes during meiosis. This process leads to the generation of genetic diversity and can result in the formation of new gene variants through mechanisms such as duplications, deletions, and exon shuffling.
- Introns also play a role in alternative splicing. The RNA polymerase synthesizes a pre-mRNA transcript, which includes both introns and exons. However, introns do not code for proteins and need to be removed before translation can occur. Through alternative splicing, different combinations of exons can be assembled, allowing a single gene to generate multiple protein isoforms. This increases the diversity and complexity of the proteome.
- During splicing, specific sequences within introns, known as spliceosome recognition sites, help the spliceosome machinery identify the boundaries between introns and exons. The spliceosome is composed of small nuclear ribonucleoproteins (snRNPs) that recognize these recognition sites. Several snRNPs combine to form the spliceosome, which carries out the splicing process in three steps.
- In summary, introns have important functions in genetic regulation and diversity. They contribute to genetic variation by promoting crossover and recombination, and they enable alternative splicing, which allows for the generation of multiple protein isoforms from a single gene. Despite being initially labeled as “junk DNA,” introns have emerged as key players in the complexity and regulation of gene expression.
Structure of Introns
Introns are non-coding DNA sequences that lie between exons within genes. Unlike exons, introns do not directly contribute to the protein-coding sequence of a gene. Here are the key features of the structure of introns:
- Non-Coding Sequences: Introns consist of non-coding DNA sequences that do not contain the necessary information for protein synthesis. They are typically rich in repetitive sequences and have a higher variability compared to exons.
- Length and Size: Introns can vary significantly in length, ranging from a few dozen nucleotides to thousands of nucleotides. Some introns are relatively short, while others can be quite long, making them a major component of gene length in some cases.
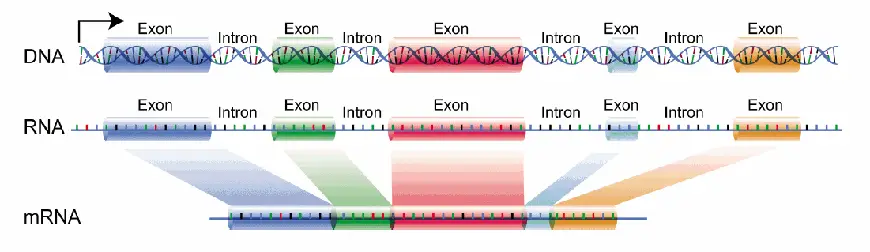
- Position within Genes: Introns are located between exons within a gene. They can be present in different positions, such as between consecutive exons or interspersed between multiple exons within a gene. The number and arrangement of introns can vary between genes and organisms.
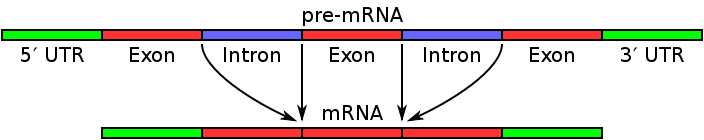
- Splice Sites: Introns contain specific nucleotide sequences at their boundaries, known as splice sites. These splice sites include the 5′ splice site and the 3′ splice site. The 5′ splice site marks the beginning of an intron, while the 3′ splice site marks the end. These splice sites are recognized by the spliceosome, a complex of RNA and protein molecules, during the process of RNA splicing.
- RNA Splicing: During RNA splicing, the introns are removed from the pre-mRNA transcript, and the exons are joined together to form a mature mRNA molecule. The excision of introns and the precise joining of exons are essential steps in the conversion of pre-mRNA into functional mRNA that can be translated into proteins.
- Alternative Splicing: Introns play a crucial role in alternative splicing, a process that allows for the production of multiple mRNA isoforms from a single gene. Alternative splicing involves the selective inclusion or exclusion of different exons and/or introns in the final mRNA transcript. This process contributes to the generation of protein diversity by producing different protein isoforms with distinct functions from a single gene.
What is Exons?
- Exons are the protein-coding DNA sequences within a gene that contain the necessary codons and information required for protein synthesis.
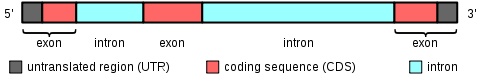
- In the genome, exons represent the expressed regions that are transcribed into mRNA and eventually translated into proteins. In eukaryotic genes, exons are interspersed with non-coding introns. During the process of RNA splicing, the introns are removed, and the exons are joined together to form mature messenger RNA (mRNA).
- The entire set of exons present in an organism’s genome is referred to as the exome. In protein-coding genes, exons encompass not only the coding sequences but also the 5′ and 3′ untranslated regions. After transcription, the resulting RNA molecule contains both exons and introns. However, during RNA splicing, the introns are excised, leaving behind the exons in the mature mRNA transcripts.
- Exons play a crucial role in protein synthesis as they contain the codons that encode the amino acid sequence of a protein. Different combinations of exons can be utilized through alternative splicing, a process that increases the diversity of proteins produced from a single gene. Alternative splicing allows for the arrangement of exons in various sequences, resulting in different protein isoforms.
- Exon shuffling is another process related to the rearrangement of exons. During recombination events, exons or sister chromosomes can be exchanged, leading to the generation of novel gene sequences and protein functions.
- The presence of exons and introns in genes allows for the phenomenon of exonization, where some introns might evolve into exons over time. This process contributes to the expansion and diversification of the coding regions within the genome.
- In the human genome, exons account for only approximately 1% of the total genome, while the majority is occupied by introns and intergenic DNA. However, the significance of exons lies in their essential role in protein coding, alternative splicing, and the generation of protein diversity.
- Overall, exons are vital elements in gene expression and protein synthesis, encoding the information required for the formation of functional proteins and contributing to the complexity and adaptability of organisms through alternative splicing and exon shuffling.
Functions of Exons
Exons serve several important functions in gene expression and protein synthesis. Here are some key functions of exons:
- Protein-coding: Exons contain the coding sequences that specify the amino acid sequence of a protein. They provide the necessary information for the translation of mRNA into functional proteins. The arrangement and order of exons within a gene determine the primary structure of the resulting protein.
- Alternative Splicing: Exons play a crucial role in alternative splicing, a process by which different combinations of exons can be included or excluded from the final mRNA transcript. Alternative splicing allows for the production of multiple mRNA isoforms from a single gene, thereby expanding the proteomic diversity. It enables the generation of different protein variants with distinct functions or tissue-specific expression patterns.
- Genetic Variation: Exons contribute to genetic variation through processes such as exon shuffling and exonization. Exon shuffling involves the exchange of exons between different genes during recombination, leading to the creation of novel gene sequences and protein functions. Exonization refers to the conversion of intronic regions into exons, expanding the coding potential of the genome.
- Regulation of Gene Expression: Exons can contain regulatory elements that influence gene expression. They may include enhancers or silencers that interact with transcription factors or other regulatory molecules to modulate the level of gene expression. By affecting the stability, localization, or translational efficiency of mRNA, exons can contribute to the fine-tuning of gene regulation.
- Protein Domains and Structure: Exons often correspond to functional domains within proteins. Different exons can encode specific structural motifs, functional domains, or protein interaction sites. The combination and arrangement of exons contribute to the overall three-dimensional structure and function of the protein.
- Evolutionary Conservation: Exons tend to be more conserved across species compared to intronic regions. Highly conserved exons are often associated with essential cellular processes or core biological functions. The conservation of exons reflects their functional significance and the selective pressure to maintain their sequences over evolutionary time.
Structure of Exons
Exons are the coding regions within genes that contain the necessary information for protein synthesis. The structure of exons is characterized by specific features and arrangements. Here are the key aspects of the structure of exons:
- Coding Sequences: Exons contain the DNA sequences that encode the amino acid sequence of a protein. These coding sequences are composed of triplets of nucleotides called codons. Each codon specifies a particular amino acid, and the sequential arrangement of codons determines the primary structure of the protein.
- Start and Stop Codons: Exons typically begin with a start codon, which is usually AUG, and they end with one or more stop codons, such as UAA, UAG, or UGA. The start codon initiates translation, and the stop codon signals the termination of protein synthesis.
- Splice Sites: Exons are connected to each other by splice sites, which are located at the boundaries between exons and introns. The splice sites consist of specific nucleotide sequences that are recognized by the spliceosome, a complex of RNA and protein molecules involved in the process of RNA splicing. The spliceosome removes introns and joins the exons together to form a mature mRNA transcript.
- Alternative Splicing Variants: Exons can be alternatively spliced, meaning that different combinations of exons can be included or excluded in the final mRNA transcript. This alternative splicing process gives rise to multiple mRNA isoforms and allows for the production of different protein variants from a single gene. The presence of alternative splice sites within exons contributes to the generation of protein diversity.
- Exon-Intron Junctions: Exon sequences are interrupted by introns, which are non-coding regions of DNA. At the exon-intron junctions, specific nucleotide sequences, such as the 5′ splice site and the 3′ splice site, mark the boundaries between exons and introns. These sequences are recognized by the spliceosome during RNA splicing, enabling the accurate removal of introns and the proper joining of exons.
- Exon Size and Number: Exon size can vary significantly, ranging from a few nucleotides to several thousand nucleotides. Some exons are relatively short, encoding only a small portion of the protein, while others may span a considerable length and contain multiple functional domains. Genes can have different numbers of exons, depending on their complexity. Some genes consist of a single exon, while others may contain dozens or even hundreds of exons.
Difference Between Introns and Exons – Introns vs Exons
| Topics | Introns | Exons |
|---|---|---|
| Definition | Introns are non-coding DNA sequences within a gene that are removed by RNA splicing during maturation of the RNA product. | Exons are protein-coding DNA sequences that contain the necessary codons or information for protein synthesis. |
| Type of sequence | Introns are non-coding sequences that do not code for any protein. | Exons are protein-coding sequences that code for specific proteins. |
| Location in the DNA | Introns are present between two exons in a DNA sequence. | Exons are located between either untranslated regions or two introns. |
| Distribution | Introns are found only in eukaryotic genomes. | Exons are found in both eukaryotic and prokaryotic genomes. |
| Location in the cell | Introns remain in the nucleus after being spliced out from the mRNA transcript during RNA processing. | Exons leave the nucleus and reach the cytoplasm after mature mRNA synthesis. |
| Present in | Introns are present in the DNA and mRNA transcripts but are not present in mature mRNA. | Exons are present in DNA, mRNA transcripts, and mature RNAs. |
| Conservation | Introns exhibit varying levels of conservation, and some introns can convert into exons through the process of exonization. | Exons are highly conserved in their sequences. |
| Involvement in protein synthesis | Introns are not involved in protein synthesis. | Exons are directly involved in protein synthesis. |
| Quantity | The nuclear genome contains more introns than exons. | The nuclear genome contains fewer exons compared to introns. |
| Human genome | Approximately 24% of the human genome is composed of introns. | Only about 1% of the human genome is composed of exons. |
| Alternative splicing | Introns are removed by alternative splicing. | Alternative splicing connects two or more exons. |
| Novel gene formation | Introns can contribute to the formation of novel genes through continuous evolutionary processes. | Exons can combine in different configurations, resulting in different protein-coding sequences. |
| During RNA Splicing | Introns are removed. | Exons are joined together to form mature mRNA. |
| Mature mRNA | Introns do not contribute to the formation of mature mRNA. | Mature mRNA forms from the complete set of exons of a gene. |
| Nature of Sequences | Introns have less conserved sequences over time. | Exons have highly conserved sequences over time between species. |
| Presence in Final RNA molecule | Introns do not appear in the final RNA molecule. | Exons appear in the final RNA molecule since they possess the genetic code. |
| Importance in Protein Synthesis | Introns are not immediately important for protein synthesis since they are non-coding. | Coding sequences (exons) are of utmost importance for protein synthesis. |
| Presence in Prokaryotes and Eukaryotes | Introns are not present in prokaryotes. | Exons are present in both prokaryotes and eukaryotes. |
FAQ
What are introns and exons?
Introns are non-coding sequences within a gene that are removed during RNA splicing, while exons are the coding sequences that contain the necessary information for protein synthesis.
How do introns and exons differ in terms of their function?
Introns do not directly code for proteins and are involved in various regulatory functions, including alternative splicing and gene regulation. Exons, on the other hand, contain the coding sequences that are translated into proteins.
Where are introns and exons located within genes?
Introns are located between exons within a gene sequence.
Do introns and exons exist in both prokaryotes and eukaryotes?
Introns are mainly found in eukaryotic genomes, while exons are present in both prokaryotic and eukaryotic genomes.
How are introns and exons involved in the process of RNA splicing?
During RNA splicing, introns are removed from the pre-mRNA molecule, and the remaining exons are joined together to form a mature mRNA transcript that can be translated into protein.
What is alternative splicing, and how does it relate to introns and exons?
Alternative splicing is a process where different combinations of exons within a gene can be included or excluded in the final mRNA transcript. This process allows a single gene to produce multiple protein isoforms, increasing the diversity of proteins generated from a single gene.
Are introns and exons conserved across different species?
Exons are highly conserved across species because they contain the coding sequences necessary for protein synthesis. Introns, on the other hand, show lower conservation and can vary in length and sequence between different species.
What is the significance of introns and exons in protein synthesis?
Exons contain the information required for protein synthesis, while introns play important roles in gene regulation, alternative splicing, and the generation of protein diversity.
Can introns convert into exons or vice versa?
In rare cases, introns can undergo a process called exonization, where they acquire the necessary elements to become exons and code for proteins. However, this is a relatively rare occurrence.
How do introns and exons contribute to genetic variation and evolution?
Introns can increase genetic variation through processes such as exon shuffling, where exons can be exchanged between different genes during recombination. This can lead to the creation of new gene variants. Additionally, alternative splicing, influenced by introns and exons, contributes to the diversity of proteins and can play a role in evolutionary adaptation.
it is well understandable contents