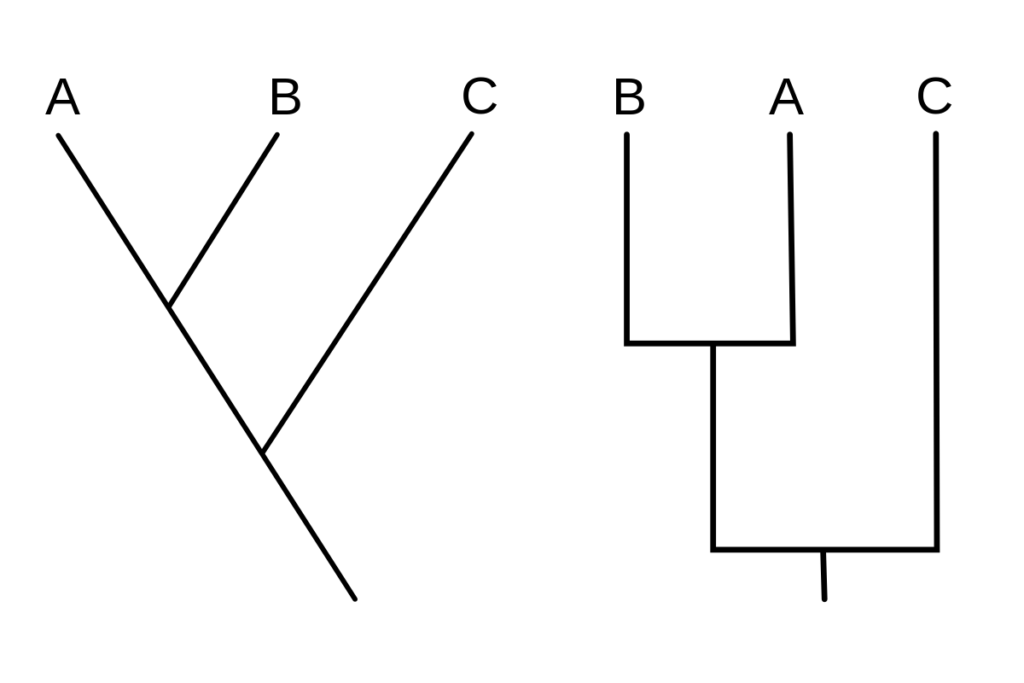
What is Cladogram tree?
- A cladogram is a form of tree diagram that depicts the evolutionary relationships between a group of organisms on the basis of shared characteristics or traits. It is a visual representation of cladistic analysis, a method of phylogenetic analysis that identifies and groups organisms based on their shared derived characteristics.
- The branching patterns in a cladogram represent the hypothesized evolutionary relationships between the organisms. Nodes represent common ancestors, while branches represent lineages or evolutionary pathways. Typically, the length of the branches is arbitrary and does not indicate evolutionary time or degree of divergence.
- Cladogram nodes represent points where novel characteristics or traits evolved. These characteristics are referred to as derived traits or synapomorphies, which are traits that are shared by some group members but not by those outside the group.
- Cladograms are frequently used to illustrate the relationships between species or groups of organisms, and they are especially useful for illustrating patterns of common ancestry and evolutionary relatedness. They provide a visual representation of the evolutionary history and can be used to infer the order and timeline of evolutionary events.
- Importantly, cladograms represent a specific evolutionary hypothesis and are subject to revision and refinement as new data and analyses become available.
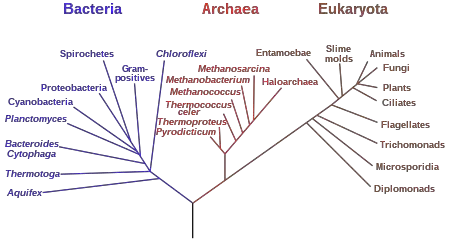
What is Phylogenetic tree?
- A phylogenetic tree, also referred to as an evolutionary tree or a tree of life, is a branching diagram that depicts the evolutionary relationships between various organisms or groups of organisms. It illustrates the pattern of descent and evolutionary history of taxonomic units or species from a common ancestor.
- The branching patterns of a phylogenetic tree illustrate the relationships between organisms. The length of the branches often indicates the amount of evolutionary change or the amount of time that has passed since divergence. A speciation event occurred when a common ancestor gave rise to two distinct lineages at the point where two branches divided apart.
- Evidence from various sources, such as genetic data (DNA or protein sequences), morphological characteristics, or a combination of both, is used to construct phylogenetic trees. By comparing and analyzing these shared characteristics, scientists can infer the evolutionary relationships and sequence of events.
- The structure of a phylogenetic tree indicates the degree to which organisms are related. Species with more recent common antecedents are closer together on the evolutionary tree than those that diverged earlier in evolutionary history. The ends of the branches, known as terminal taxa, represent the analyzed species or groups.
- Phylogenetic trees are a graphical representation of evolutionary relationships, illustrative of branching patterns and hierarchical organization of life. They are essential instruments in evolutionary biology, systematics, and comparative genomics, aiding in the comprehension of the evolution of organisms, their diversification, and their classification.
- Importantly, phylogenetic trees are founded on scientific inference and are subject to revision and refinement as new data and analytical techniques emerge. They serve as working hypotheses that are continually evaluated and revised to enhance our knowledge of the evolutionary relationships between organisms.
Cladogram vs Phylogenetic tree
| Cladogram | Phylogenetic Tree |
|---|---|
| Represents evolutionary relationships based on shared derived traits (synapomorphies) | Represents evolutionary relationships among organisms or groups of organisms |
| Focuses on identifying and grouping organisms based on shared characteristics | Represents the pattern of descent and the evolutionary history of species or taxonomic units |
| Branching patterns illustrate the relationships between organisms | Branching patterns illustrate the relationships between organisms |
| Branch lengths do not typically represent evolutionary time or degree of divergence | Branch lengths may represent evolutionary time or degree of divergence |
| Nodes represent points where new characteristics or traits evolved | Nodes represent common ancestors |
| Used in cladistic analysis, which emphasizes shared derived traits | Used in phylogenetic analysis, which considers various data types (genetic, morphological) |
| Can be simpler in structure, focusing on specific traits or groups | Can be more complex, encompassing a broader range of organisms and traits |
| Can be created based on a single trait or a limited set of traits | Requires comprehensive data and analysis, often involving multiple traits and species |
| Subject to revision and refinement as new data and analysis become available | Subject to revision and refinement as new data and analysis become available |
FAQ
What is a cladogram?
A cladogram is a diagram that represents evolutionary relationships based on shared derived traits among organisms. It shows the pattern of evolutionary branching and the grouping of organisms into nested clades.
How is a cladogram different from a phylogenetic tree?
A cladogram is a type of phylogenetic tree that specifically focuses on shared derived traits. While all cladograms are phylogenetic trees, not all phylogenetic trees are cladograms, as the latter specifically emphasize shared characteristics.
What are shared derived traits?
Shared derived traits, also known as synapomorphies, are characteristics or traits that are shared among a group of organisms and are derived from a common ancestor. These traits are used to determine the evolutionary relationships and groupings within a cladogram.
How are cladograms constructed?
Cladograms are constructed by analyzing the presence or absence of shared derived traits among different organisms. These traits are used to define nested groups or clades, and the relationships among the groups are depicted by branching patterns.
Are cladograms based solely on genetic data?
Cladograms can be constructed using various types of data, including genetic sequences, morphological traits, or a combination of both. The choice of data depends on the available information and the research objectives.
Can a cladogram show the timing of evolutionary events?
Cladograms generally do not represent the timing of evolutionary events. They focus on illustrating the pattern of relationships and the branching order of organisms, rather than indicating the precise timing of evolutionary changes.
How do you interpret a cladogram?
To interpret a cladogram, one examines the branching patterns, identifies common ancestors at nodes, and analyzes the relationships between different groups of organisms. Closer branching points indicate more recent common ancestry.
What is the difference between a node and a branch in a cladogram?
In a cladogram, a node represents a common ancestor from which two or more branches emerge. Nodes indicate points of evolutionary divergence. The branches represent lineages or evolutionary pathways leading to different groups of organisms.
Can cladograms change over time?
Yes, cladograms are subject to change and revision as new data, improved analytical methods, or alternative interpretations become available. The understanding of evolutionary relationships continually evolves, leading to refinements in cladogram construction.
How are cladograms used in evolutionary biology?
Cladograms are used in evolutionary biology to understand the relationships among organisms, trace the pattern of evolution, classify organisms into groups, reconstruct ancestral characteristics, and make inferences about common ancestry and evolutionary history.